- Department of Biochemistry and Molecular Biology, Faculty of Veterinary Medicine, Wrocław University of Environmental and Life Sciences, Wrocław, Poland
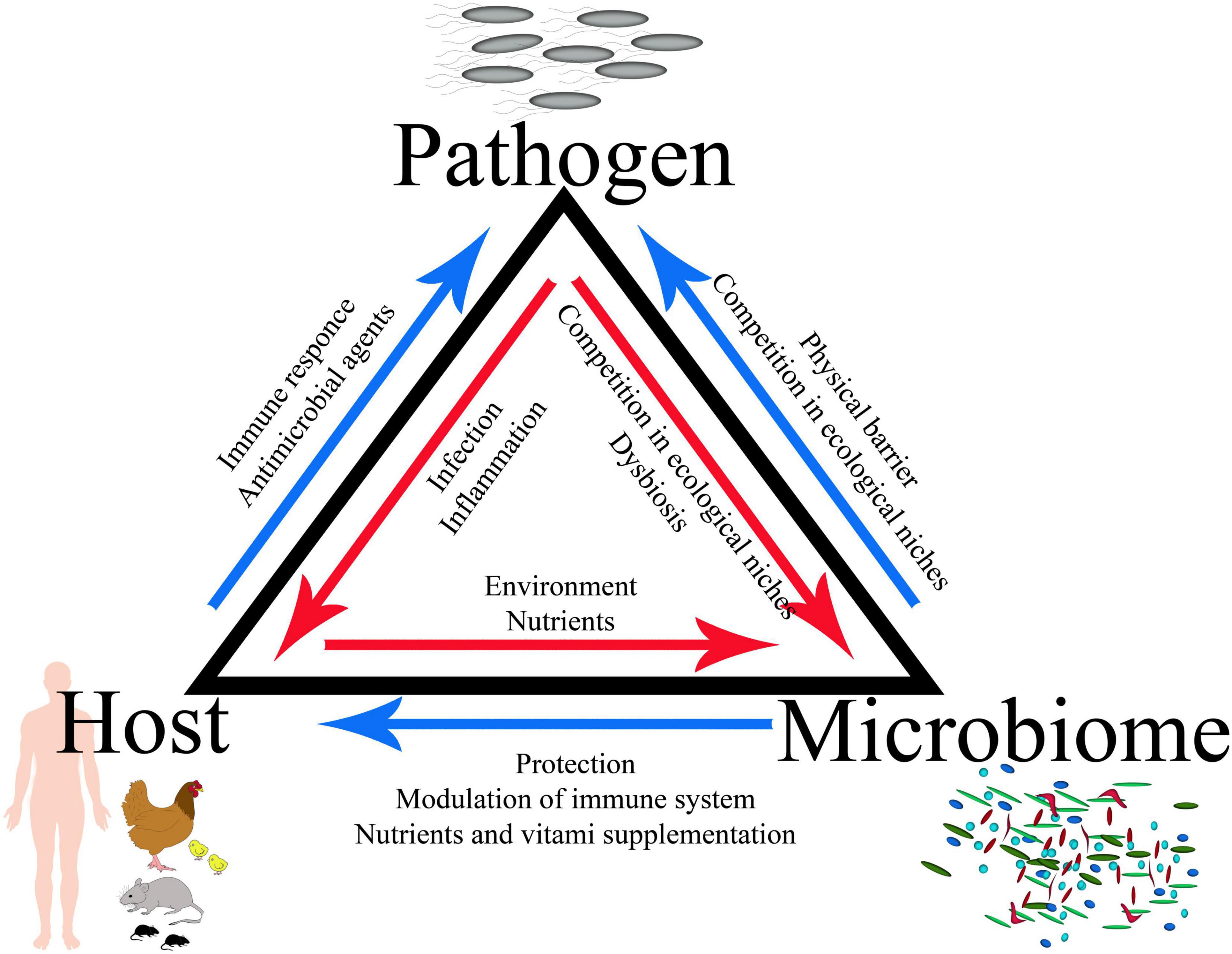
Colonization of the gastrointestinal (GI) tract by enteric pathogens occurs in a context strongly determined by host-specific gut microbiota, which can significantly affect the outcome of infection. The complex gameplay between the trillions of microbes that inhabit the GI tract, the host, and the infecting pathogen defines a specific triangle of interaction; therefore, a complete model of infection should consider all of these elements. Many different infection models have been developed to explain the complexity of these interactions. This review sheds light on current knowledge, along with the strengths and limitations of in vitro and in vivo models utilized in the study of Salmonella–host–microbiome interactions. These models range from the simplest experiment simulating environmental conditions using dedicated growth media through in vitro interaction with cell lines and 3-D organoid structure, and sophisticated “gut on a chip” systems, ending in various animal models. Finally, the challenges facing this field of research and the important future directions are outlined.
Key Messages
• The GI tract is occupied by trillions of microbes; therefore, a complete model of infection should include interactions between the host, its microbiota, and the infecting pathogen, which defines a specific triangle of interaction.
• Most of the currently used infection models may be applied to investigate the host–Salmonella–microbiota triangle.
• Appropriate model selection may shed new light on Salmonella infection.
Introduction
Salmonella is a Gram-negative foodborne pathogen with the ability to infect a wide range of species. Depending on the serovar and infected host, Salmonella can cause diseases with different clinical symptoms, ranging from alimentary tract disturbances (gastroenteritis) caused by host-unrestricted serovars such as Enteritidis or Typhimurium, to invasive typhoid-like diseases caused by host-restricted serovars, such as Typhi or Gallinarum (Uzzau et al., 2000). To establish successful infection, enteric pathogens, such as Salmonella, must sense and respond to newly encountered host environments to regulate the expression of critical virulence factors, such as flagella, fimbriae, invasins, and secretion systems present in Salmonella Pathogenicity Islands (SPI) to actively reach, attach, and invade host cells (Saini et al., 2009; de Masi et al., 2017; Kolenda et al., 2019). Interestingly, allelic variations of those virulence factors are often directly related to the phenomena of host specificity (Grzymajło et al., 2013; Yue et al., 2015; de Masi et al., 2017). After adhering to the host cell surface, Salmonella actively invades and survives intracellularly in phagocytic and non-phagocytic cells (Malik-Kale et al., 2011). Most studies on the early stages of Salmonella infection have focused on a direct interaction with host cells, such as enterocytes, M-cells, or macrophages, in many cases with the use of immortalized cell line models (Brosnahan and Brown, 2012; Torraca et al., 2014; Kolenda et al., 2019). However, colonization of the GI tract by enteric pathogens always occurs in a broader context, strongly determined by host-specific gut microbiota, which can heavily affect host–pathogen interactions. The gut microbiome appears to be one of the critical factors in resistance to enteric pathogen colonization; however, the exact mechanism has not been fully elucidated (Velazquez et al., 2019; Rogers et al., 2021).
The GI tract is occupied by trillions of microbes, including complicated bacterial, fungal, and viral populations; therefore, a complete infection model should include the interactions between the host, its microbiota, and the infecting pathogen, which defines a specific triangle of interaction (Figures 1, 2). Bacteria account for around 93% of the unique gene repertoire of the microbiome, viruses less than 6%, and fungi around 0.1% (Shkoporov and Hill, 2019). Despite recent research demonstrating the important role of viral and fungal microbiota (Lecuit and Eloit, 2017; Pérez, 2021; Thielemann et al., 2022), this review will focus on the largest part of the microbiome—bacteria.
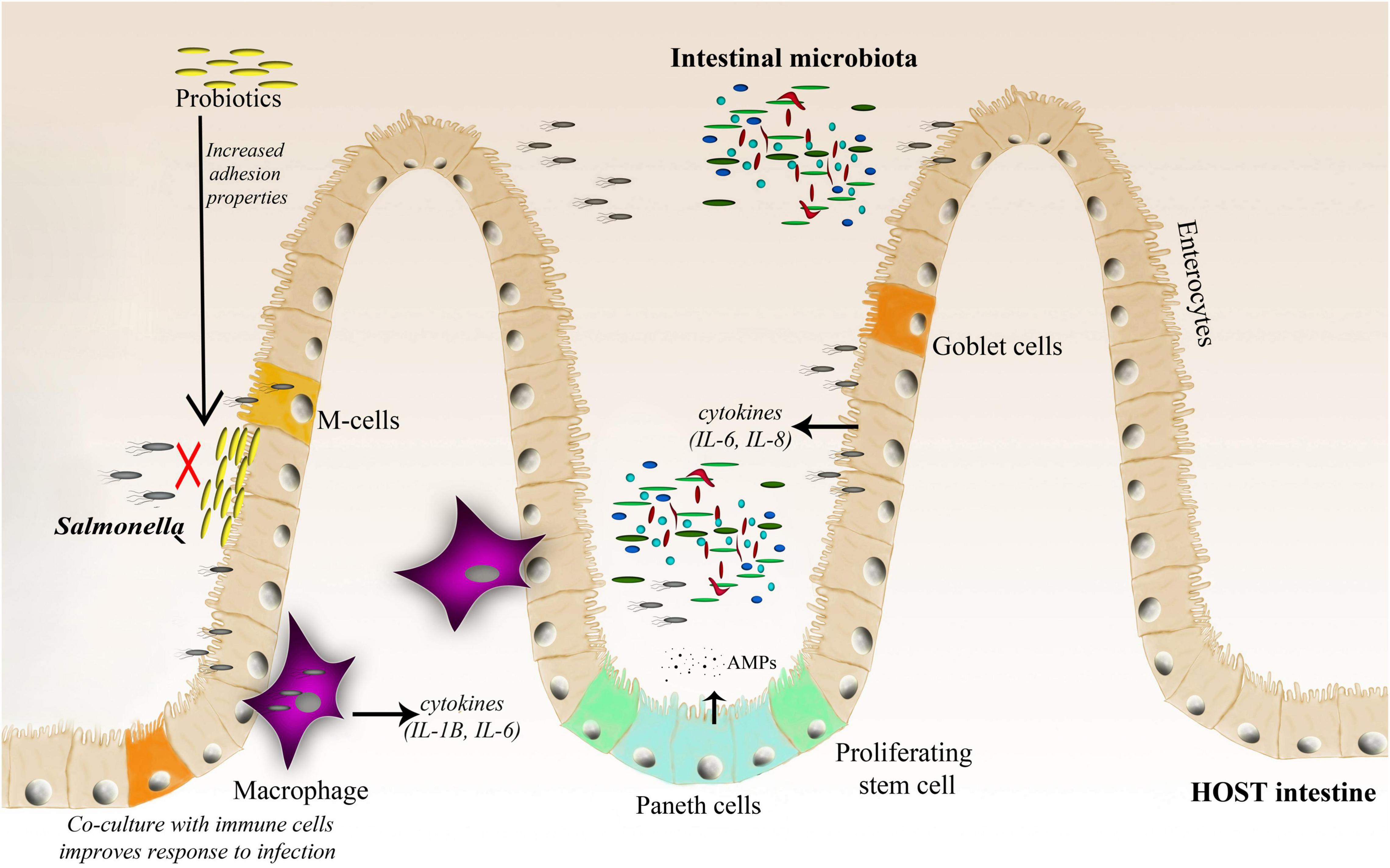
Figure 2. A schematic representation of the Salmonella–host–microbiota interaction. This figure highlights various factors (immune response, microbiota adhesion, and microbiota competition with the invading pathogen) affecting the gut microbiota composition and infection outcome.
Bacteroidetes and Firmicutes dominate the human gut, accounting for up to 90% of the bacteria in the adult human gut (Jandhyala et al., 2015), however, their composition changes throughout the life. Among the trillions of microorganisms that inhabit the human intestine, there are more than 1,000 species, at least 160 bacterial species per person, and 100 times more genes than in the human genome (Turnbaugh et al., 2007; Gahan et al., 2011). The complexity and diversity of the microbiome are determined by multiple factors, such as age, genetics, diet, pharmacological treatments, and general lifestyle (Vasquez et al., 2018; Zhang et al., 2018). There are two major features of microbial communities that are postulated to mediate resistance to pathogens: community species richness and community composition. A healthy and balanced intestinal microbiome provides many benefits to the host, such as proper development of the immune system, stimulation of proper intestinal tract cell development (Salzman et al., 2010), absorption of nutrients, support for vitamin production, and finally protection against pathogenic infections (Huttenhower et al., 2012). Disruption of the microbiota (known as dysbiosis) due to various factors, such as antibiotic treatment or an unbalanced diet, favors infection by different pathogens, including Salmonella. Gut microbes promote colonization resistance by competing with pathogens for nutrients, priming, modulating the host immune system, and directly targeting other microbes with metabolites (Vasquez et al., 2018). Furthermore, the microbiota can act as a physical barrier against invading bacteria by blocking pathogen access to the epithelial layer (Sassone-Corsi and Raffatellu, 2015), and actively stimulates Paneth cells for the production of antimicrobial peptides (AMPs) (Salzman et al., 2007; Yokoi et al., 2019; Figure 2).
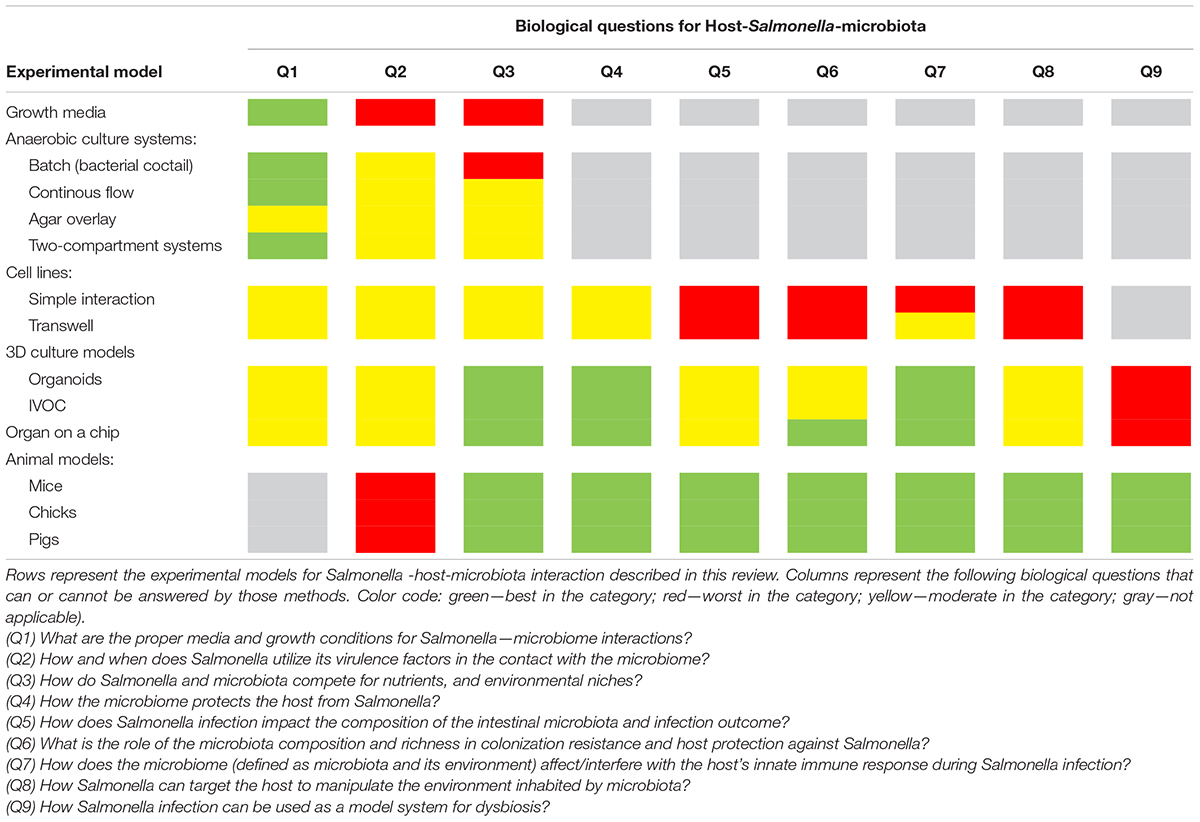
Despite numerous studies, there are still many unknowns about the effect of perturbations in the intestinal microbiota on host susceptibility to invading pathogens. Enteric pathogens, such as Salmonella, can interact intensively with the intestinal microbiota, thereby affecting the composition of microbiome, which changes the outcome of the infection (Sekirov et al., 2008; Bucci et al., 2016). Several unanswered questions have arisen over the years about Salmonella–host–microbiome interactions (Table 1):
Q1) What are the proper media and growth conditions for Salmonella–microbiome interactions?
Q2) How and when does Salmonella use its virulence factors in contact with the microbiome?
Q3) How do Salmonella and microbiota compete for nutrients and environmental niches?
Q4) How does the microbiome protect the host from Salmonella?
Q5) How does Salmonella infection impact the composition of the intestinal microbiota and the outcome of infection?
Q6) What is the role of the microbiota composition and richness in colonization resistance and host protection against Salmonella?
Q7) How does the microbiome (defined as microbiota and its environment) affect/interfere with the host innate immune response during Salmonella infection?
Q8) How can Salmonella target the host to manipulate the environment inhabited by the microbiota?
Q9) How can Salmonella infection be used as a model system for dysbiosis?
Many different infection models have been developed to address these questions and to understand this three-way gameplay between the pathogen, host, and microbiota. This review addresses the current knowledge, along with the strengths and limitations of in vitro and in vivo models used in the study of Salmonella–host–microbiome interactions.
Models for Host–Pathogen–Microbiome Research in Salmonella Studies
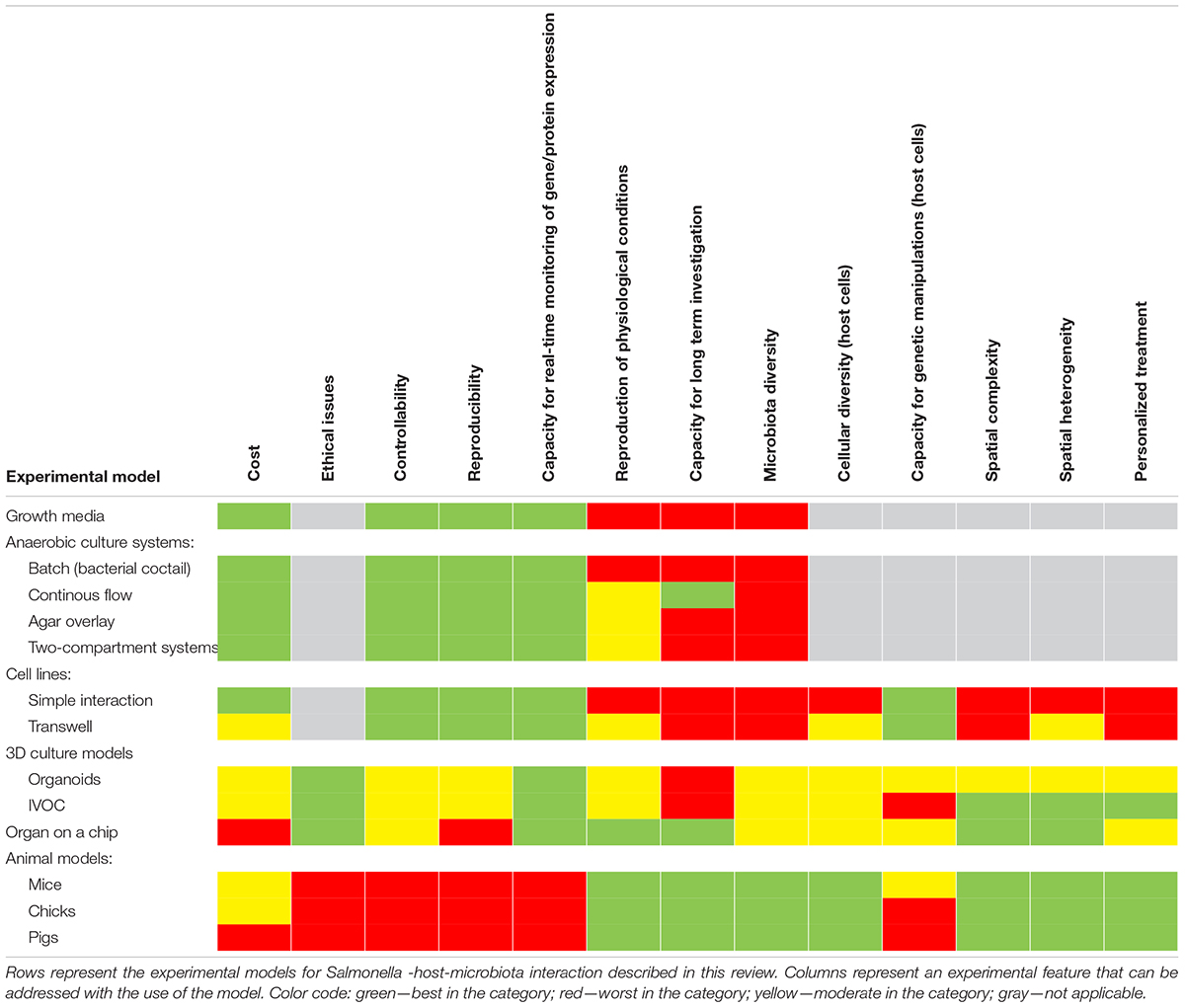
Many experimental models have been developed to study Salmonella infections, ranging from in vitro, ex vivo, and finally in vivo studies (Table 2 and Figure 3). Most of them have been successfully applied to study Salmonella interactions with the microbiota or the host–Salmonella–microbiota triangle. In vitro experiments focusing on environmental cues and stimuli that mimic the gut environment are less expensive, simpler, and can be a good alternative to more complicated models, especially in the screening phase (Vartoukian et al., 2010). When more complex interaction patterns are considered, established intestinal cell lines can serve as a model for host–pathogen–microbiota interactions during the early stages of bacterial pathogenesis (Brosnahan and Brown, 2012; Carey and Kostrzynska, 2013). Immortalized cell lines can serve as a good and cost-effective model for the early host–microbiota screening experiments; however, due to their relative lack of complexity, the sophisticated architecture of the intestine cannot be fully simulated. To mimic the complex organization of intestinal epithelia, a three-dimensional (3D) organotypic model (Höner zu Bentrup et al., 2006) and an in vitro organ culture (IVOC) model (Fang et al., 2013) have been developed to better represent the characteristics associated with intestinal epithelia in vivo. In contrast to two-dimensional (2D) cell cultures, the 3D organotypic model has a better organization of junctional, extracellular matrix, brush-border proteins, Paneth cells secreting AMPs, and highly localized mucin production (Höner zu Bentrup et al., 2006; Yin and Zhou, 2018; Gieryńska et al., 2022). In addition, its ability to co-culture with immune cells, such as macrophages, enables the formation of a more physiologically relevant intestinal model, which is more appropriate to study host–bacteria interactions in more detail and the immune responses caused by the microbial, probiotic, and pathogenic infection. On the microbial side, organoids and enteroids have been used for host–pathogen and host–commensal studies (Aoki-Yoshida et al., 2016; Hill and Spence, 2017; Co et al., 2019). In vitro experiments are key to understanding the mechanisms of infection in the context of microbiota; however, it is difficult to extrapolate these results to the animal intestinal tract where other factors, such as peristaltic movement and complete host defense system, could interfere with the process. Moreover, it is not an easy task to transplant the complex microbiota communities living in the gut to a relevant in vitro system, mostly because of its limited viability and high sensitivity to oxygen. Therefore, animal models are frequently used to explore the course and mechanisms of Salmonella infections in the context of their interactions with the microbiota.
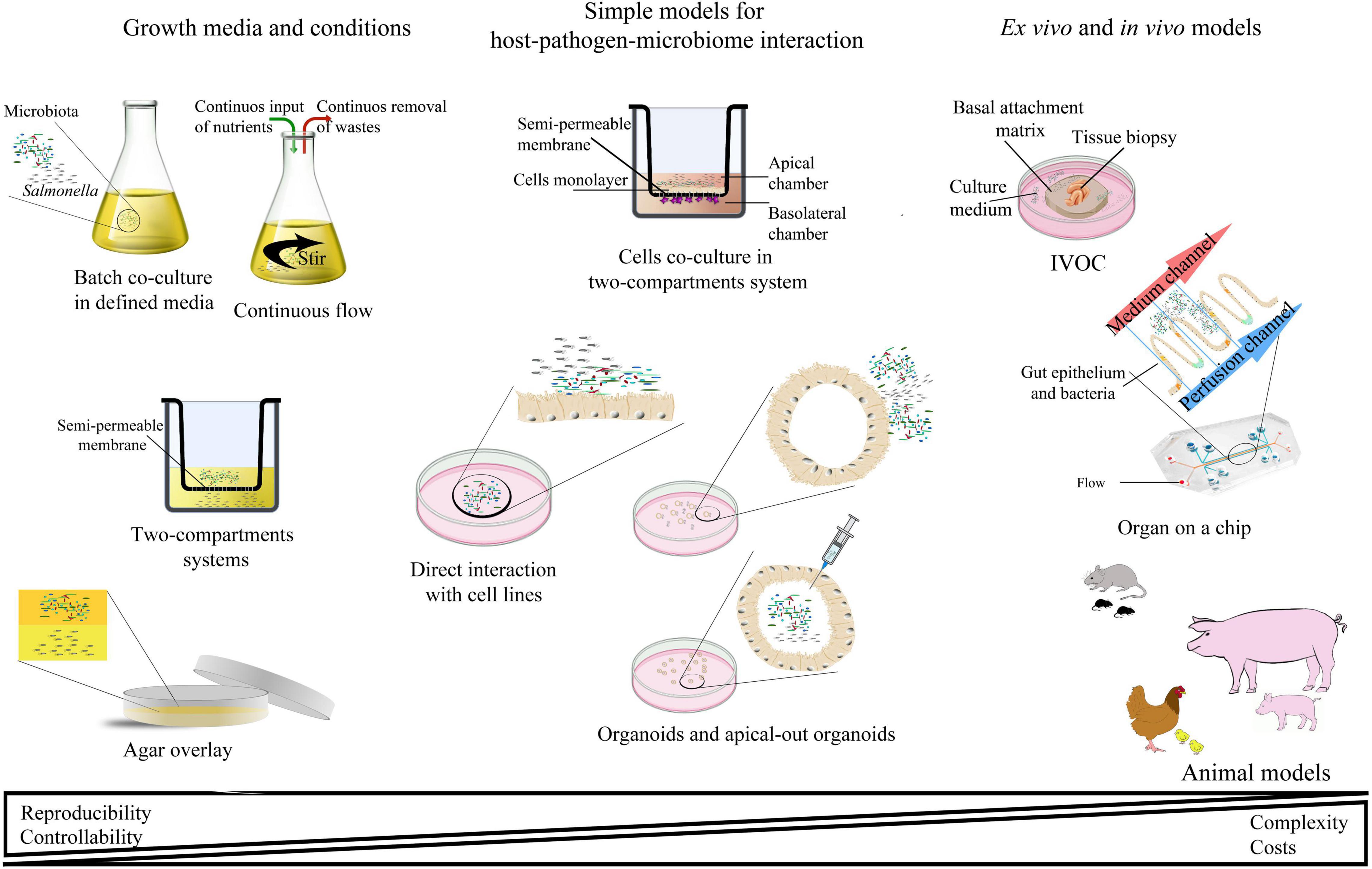
Figure 3. Schematic representation of the most abundant interaction models for studying host–pathogen–microbiota interactions.
In deciding which model should be applied to answer experimental questions, one needs to find a balance between simple systems, which paint a limited picture of the interaction but are easy-to-maintain and control, and highly complex models that fulfill many different aspects of the interaction, but are more difficult to control and utilize (Table 2 and Figure 3). The incorporation of a third player, a complex microbiome population, generates additional limitations and challenges; however, it leads to the generation of a big picture of pathogenic infection.
Growth Media and Anaerobic Culture Systems
In vitro studies that simulate the gut environment are useful for investigating the mechanisms of pathogen–microbiota interactions by mimicking the host milieu. These experiments are less expensive, simpler, provide better control, enable easier manipulation, have the potential to find appropriate conditions to analyze specific bacterial interactions, and, finally, require no ethical restrictions (Table 1). However, such in vitro experiments also have disadvantages. For example, among the microbiome, many bacterial species are considered uncultivable (Eckburg et al., 2005; Vartoukian et al., 2010). This last statement is only partially true as most microorganisms are amenable to cultivation under appropriate culture conditions. Some anaerobes can survive in oxygen-rich conditions by forming spores (Kato et al., 1997). Others, such as many gut-inhibiting species, are obligate anaerobes and can be cultured only in oxygen-free conditions (Vartoukian et al., 2010).
Even the selection of growth media suitable for complex microbiota cannot ensure that these conditions are suitable for enteric pathogens, such as Salmonella. For example, selenite-rich media enhance the growth of Salmonella species but inhibit fecal streptococci (Ito et al., 2019). Growth media need to be modified to obtain an appropriate concentration of proteins, sugars, and/or salts to mimic conditions in the ileum or colon. Many other features, such as pH, temperature, oxygen level, and different energy sources, affect media selectivity. Correctly selected media allow the growth and detection of less abundant bacteria, which may be missed in culture-independent studies. However, despite experiments with different culture media and conditions, microbiota composition from fecal inoculum differs in growth reactor cultures due to changes in the environment, such as medium composition, pH, or retention time (Cleusix et al., 2008; Ma et al., 2010; Naimi et al., 2020).
There are several strategies to provide appropriate media and growth conditions for microbiota-Salmonella interactions. The easiest solution is to measure how the selected environment affects both sides of the interaction: microbiota composition and richness, and the Salmonella proliferation and gene expression profile. Frequently, bacteria recovered from fecal samples are diluted in phosphate buffer and inoculated with selected media. Initially, commercial non-selective media, such as 5% sheep blood agar, M9, gut microbiota medium (GMM), and gut anaerobic medium (GAM), were used. The microbiota were grown under a range of oxygen availability (aerobic conditions, aerobic conditions with 2.5% CO2 or 5% CO2, microaerophilic conditions, and anaerobic conditions) at different temperatures and with different incubation times (from hours to months) (Rettedal et al., 2014). Subsequently, specific media were suggested for in vitro interactions between Salmonella and the microbiota. For example, Saccharomyces cerevisiae fermentation product (SCFP) supplemented with XPC (Original XPC™ Diamond V, Cedar Rapids, IA, United States) significantly reduced the population of Salmonella Typhimurium, suggesting that XPC enhances the inhibition of Salmonella growth by the chicken cecal microbiota (Roto et al., 2015; Park et al., 2017).
With the exception of dedicated media, there is still a need for systems that can closely mimic the in vivo situation, and therefore reproduce the physiological parameters of the GI environment. To date, several in vitro culture methods have been applied to study the interaction between the intestinal microbiota and invading pathogens, including Salmonella. The simplest model is described as a “bacterial cocktail” in which defined GI microbiota and invading pathogens are mixed in one compartment. Batch cultures that mimic the conditions of the intestine are frequently used as models of the impact of microbiota on Salmonella growth. For example, batch inoculation of fresh fecal bacteria with S. Typhimurium severely affects pathogen survival (Levine et al., 2012; Avendaño-Pérez et al., 2015). Similar systems were used to investigate the capacity to inhibit S. Typhimurium growth by 973 anaerobic bacterial culture supernatants and 16 species isolated from pig feces (Levine et al., 2012). Researchers connected this 1,000 up to 100,000-fold Salmonella growth inhibition with the production of fermentation acids during anaerobic growth; therefore, the pH was reduced to 5 or less. Decreased pH caused by probiotic Lactobacillus strains as a reason for Salmonella growth inhibition was also confirmed in the co-incubation model (Fayol-Messaoudi et al., 2005), co-culture models (Adetoye et al., 2018; Burkholder et al., 2019), and by a modified agar overlay method (Adetoye et al., 2018). In the last one, agar plates incubated under microaerophilic conditions were further covered with Salmonella diluted in soft agar [Mueller Hinton (MH) soft agar (0.7% agar)], followed by measuring the size of the growth inhibition zone.
Another frequently used system for pathogen–microbiota interaction, a continuous flow model, brings substantial advantages to the topic. First, it simulates transit in the intestine, and second, it supports fresh substrate supplementation and the removal of toxic products (Ushijima and Seto, 1991; Avendaño-Pérez et al., 2015; Fehlbaum et al., 2015; Poeker et al., 2019). The continuous culture model for Salmonella–microbiota interaction was based on the original Macfarlane model (Macfarlane et al., 1998) inoculated with diluted feces. Despite its limited microbial stability, this system was used in experiments with enteropathogens. Initially, exogenous bacteria were washed out of the system (Carman and Woodburn, 2001; Payne et al., 2003). Modification of this model, with fecal microbiota immobilized on gel beads in continuous-flow anaerobic cultures (Cinquin et al., 2006a,b) was successfully applied for further investigations with Salmonella (Le Blay et al., 2009; Dostal et al., 2014). Researchers suggest that the immobilization of Salmonella on polysaccharide beads allows maintenance of pathogens even for long incubation periods, up to 43 days. Another proposal was to immobilize fecal microbiota instead of pathogens to prevent instability of the microbiome diversity. These Poly-FermS results with high stability and reproducibility were used for microbiota–pathogen interactions (Zihler Berner et al., 2013), including studies on Salmonella–microbiota interactions (Fang et al., 2013).
In addition to direct cell–cell interactions, the impact of cell-free microbiota supernatant on Salmonella growth and proliferation can be measured. Its antibacterial activity is determined either by direct incubation with the membrane filtration-sterilized supernatant or by using two-compartment systems. In the former case, Lactobacillus plantarum supernatant added to selected foodborne pathogens, including Salmonella Paratyphi A and S. Typhimurium SA2093 in serial dilutions, was shown to result in anti-Salmonella activity (Uraipan et al., 2014). Other studies have shown that heat-inactivated cell-free supernatant (CFS) of selected probiotics reduces the cytotoxicity of S. Typhimurium and inhibits its growth by approximately 50%. Moreover, the expression of the IL-8 gene initially induced by Salmonella was also reduced (Kawarizadeh et al., 2020).
The second method involves two-compartment systems, in which two bacterial populations are separated by a permeable or semi-permeable membrane and share only the growth medium. A high density of Escherichia coli was shown to inhibit S. Typhimurium growth in such settings (Avendaño-Pérez et al., 2015). In addition, the high-density population of Salmonella may inhibit microbiota growth, as S. Typhimurium inhibits Lactobacillus gasseri and Bifidobacterium bifidum growth in the other compartment (Avendaño-Pérez et al., 2015). On the other hand, using the comparison between batch and the two-compartment systems, it was shown that the growth inhibition of S. Typhimurium depends on direct cell–cell contact rather than responding to metabolites released in the medium (Avendaño-Pérez and Pin, 2013). This study demonstrates the existence of a novel interaction scheme between the intestinal microbiota and S. Typhimurium, which requires cell contact or proximity and leads to growth inhibition or loss of the cultivability of S. Typhimurium. This phenomenon may be related to the activity of the Type 6 Secretion System (T6SS), which is used by GI pathogens to compete with the intestinal microbiota (Sana et al., 2016; Anderson et al., 2017; Wood et al., 2020).
Collectively, the selection of appropriate growth media and growth conditions allows an understanding of the mutual interactions between the pathogen and the microbiota at the basic level, including competition for nutrients, metabolites released by the microbiome, and virulence factors expressed by Salmonella during contact with other bacteria.
Cell Lines
Several studies have shown that probiotics and microbiota species can affect Salmonella growth and gene expression in vitro. Established cell lines are well-tested and relatively easy-to-maintain models for the introduction of host cells into the Salmonella–microbiota equation. Moreover, the relative simplicity of cell line models allows investigation of the molecular mechanisms of microbiota activity in this interaction. Immortalized cell lines can serve as a good and cost-effective model for early screening experiments; however, due to their lack of complexity, they cannot fully mimic the complex architecture of the intestine. The major goals of such studies are to explain the immunomodulatory effect of various probiotic strains, to investigate the expression of virulence factors in Salmonella in the presence of microbiota, and, finally, to select strains that can effectively affect Salmonella adhesion to and invasion of host cells. These experiments were mainly conducted using intestinal epithelial cells (IECs), including the cell lines of both normal tissue origin and cancerous origin from different species (see below).
The simplest model used in Salmonella–host–microbiota interactions is confluent cell monolayers of IECs, such as IPEC-J2 (Schierack et al., 2011), IEC-6, or IEC-8 (Khailova et al., 2010; Liu et al., 2010) or cells of cancerous origin, such as HT-29 and Caco-2 (Carey and Kostrzynska, 2013; Kawarizadeh et al., 2021). Cell lines of normal tissue origin are morphologically and functionally similar to primary cells, develop proper microvilli, tight junctions, and Toll-like receptors (TLRs). Moreover, its innate immune response is relatively closer to primary tissue and therefore better mimics the host physiology (Schierack et al., 2006; Brosnahan and Brown, 2012). Some cell lines, like HT29-MTX cells, have the ability to differentiate into goblet cells and secrete mucin (Lesuffleur et al., 1993; Leteurtre et al., 2004). It is worth mentioning that complete confluence of cells plays an important role in such experiments because the empty spaces between the cells provide additional niches for bacteria. On the other side, well-developed tight junctions between confluent cells may act as entry sites for Salmonella (Fattinger et al., 2020).
One of the useful approaches of the cell line model is to measure the release of pro- and anti-inflammatory molecules in response to Salmonella infection and to explain the immunomodulatory effects of various microbiota. The expression profile can be measured either based on the mRNA level via quantitative polymerase chain reaction (qPCR) or at the protein level based on released cytokines (Carey and Kostrzynska, 2013; Moshiri et al., 2017; Kanmani and Kim, 2020). Probiotic strains (microbiota) added to confluent cell monolayers before, during, or after Salmonella infection for the defined period modulate cytokine expression profiles in the infected/growth media. For example, prestimulation of the HT-29 cell line monolayer with Lactobacillus brevis, Lactobacillus curvatus, and Lactobacillus pentosus for 48 h, followed by washing steps and infection with S. Typhimurium for either 3 or 12 h, there was a general reduction of the inflammatory response (Kanmani and Kim, 2020). It is worth mentioning that the order of infection plays a role in host–Salmonella–microbiota interactions. Preincubation, co-incubation, and postincubation of probiotic strains may affect Salmonella infection in a different way. For example, only preincubation of probiotic Lactobacillus and Bifidobacterium strains with the HT-29 cell line followed by S. Typhimurium D104 infection had an immunosuppressive effect on IL-8 mRNA expression and IL-8 secretion (Carey and Kostrzynska, 2013). Furthermore, in the case of Lactobacillus kefir IM002, the anti-inflammatory effect was caused by incubation with the bacterial supernatant. The probiotic strain and its supernatant were also tested by Bermudez-Brito et al. (2013) Bifidobacterium breve co-incubated with Salmonella Typhi and dendritic cells (DCs) monolayers significantly decreased the secretion of pro-inflammatory cytokines, suggesting that whole bacteria promote anti-inflammatory effects and prevent Salmonella-induced inflammation, whereas the secreted components (CFS) exert anti-inflammatory effects in the GI tract. Kanmani and Kim (2020) revealed that in the case of Lacticaseibacillus paracasei CNCM I-4034 and DC cell, probiotic bacteria as well as CFS, activates TLR signaling and decreases the production of proinflammatory cytokines in response to S. Typhi infection.
In addition to the host cell response, the expression of Salmonella virulence genes can be measured during cell line infection in the presence of probiotic strains. Lactobacillus acidophilus, Lactobacillus rhamnosus, and Lactobacillus casei were shown to affect the expression of S. Javiana virulence genes invA, prgH, pltA, and cdtB in the presence of HT29-MTX cells at the mRNA level (Burkholder et al., 2019). Moreover, the abovementioned probiotics reduce S. Javiana invasion and limit Salmonella-induced cell damage (Burkholder et al., 2019).
Adhesion to host cells, followed by invasion, is one of the crucial steps in Salmonella pathogenicity. Attachment to host proteins and gut mucosa can be affected by various types of gut microbiota, which compete for binding sites present on host proteins (Das et al., 2013; Uraipan et al., 2014). For example, the L. plantarum KSBT 56 strain isolated from dahi chenna (a fermented milk product) co-administered or 1 h after the pathogen prevents not only Salmonella Enteritidis adhesion to the HCT-116 colon epithelial cell line, but decreases its biofilm formation ability (Das et al., 2013). Preincubation with L. plantarum CIF17AN2 isolated from healthy infant feces decreased the binding of S. Typhimurium SA2093 to HT-29 cells (Uraipan et al., 2014). In this particular case, Lactobacillus strains applied before Salmonella work more efficiently, and therefore suggest that prebiotic adhesion may block Salmonella entry sites. Bacillus coagulans also significantly reduced the binding of S. Typhimurium to HT-29 cells, and no effect was observed for the supernatant at different concentrations (Kawarizadeh et al., 2019). Interestingly, the addition of 1–4% freshly prepared fecal slurry under anaerobic conditions limited this anti-Salmonella activity, whereas supplementation with saba starch significantly decreased the number of attached Salmonella. This phenomenon was caused by a lower production of fatty acids and therefore a decrease in pH. Lower adhesion levels may thus be connected either with the direct competition of bacteria for binding spots or by chemicals released by the microbiota.
The idea that probiotics block Salmonella binding sites was confirmed using IPEC-J2, a porcine IEC line isolated from neonatal piglet mid-jejunum, widely used as a model of host–pathogen interaction and supporting the invasion of at least several Salmonella serovars (Schierack et al., 2006; Brosnahan and Brown, 2012; Klasa et al., 2020). Moreover, due to the expression of some pathogen recognition receptors reported (Crocker and Feizit, 1996; Skjolaas et al., 2007), IPEC-J2 is a good model for competition between microbiota and Salmonella for binding sites. For example, preincubation and co-incubation with E. coli Nissle 1917 (EcN) and Salmonella resulted in suppression of invasion by decreased adhesion (Schierack et al., 2011). In addition to adhesion and invasion, the presence of the microbiota in host–Salmonella interactions affects the cytotoxicity caused by pathogens in a simple co-culture model. The cytotoxicity of S. Typhimurium on HT-29 cells with 90% confluency was significantly reduced when incubated with B. coagulans in comparison to cell-free probiotic supernatant (Kawarizadeh et al., 2019).
To further investigate the impact of microbiota growth media/supernatant on Salmonella–microbiota interactions, physical separation of bacteria and cells is required. Among the many systems that separate two compartments, Transwell is frequently used in host–pathogen–microbiota interactions (Tyrer et al., 2011; Bermudez-Brito et al., 2013; Yeung et al., 2013). This system was applied to verify the protection delivered by Lactobacillus strains to the Caco-2 cell line stimulated by S. Typhimurium lipopolysaccharide (LPS) (Yeung et al., 2013). Lactobacillus was found to protect tight junctions and, therefore, epithelial barrier integrity from damage caused by LPS. Another interesting use of the system is the co-culture of two different cell lines in separate compartments (Bermudez-Brito et al., 2013). For example, Caco-2 cells cultured in the upper part and human intestinal-like DCs cultured in the lower chamber mimics the in vivo conditions under which DCs open tight junctions between epithelial cells and take up bacteria directly from the intestinal lumen (Rescigno et al., 2001; Fattinger et al., 2020). DCs react differently to probiotics and pathogens in the presence and absence of IECs; therefore, co-culture models such as the one presented above may facilitate the study of host–microbe interactions. Moreover, the transwell system provides a feasible platform for the development of novel probiotics or other agents for further animal studies and clinical trials. However, the transwell system has several disadvantages: the lack of direct cell–cell contact, problems with proper migration due to the size of the pores, or inappropriate differentiation due to weak indirect cell–cell contact (Spoetti et al., 2007; Tyrer et al., 2011). Some of these caveats can be addressed by transwell co-culture models, in which bacteria, immune cells, and epithelial cells are studied together. Finally, cell line models for Salmonella–microbiota interactions can help address questions regarding the impact of such interactions on host receptor composition, organization, or modification. For example, alterations in cell glycosylation or fucosylation profiles by Salmonella may indirectly affect gut microbiota homeostasis (Hao et al., 2020).
Taken together, the introduction of cell lines as a host representation into three-handed gameplay allows for studying direct interaction between all players. Introduction of the element of the immune system, implementation of niches created by cells tight junctions and microvilli, and well as the playground for pathogen–microbiome competition for nutrients make cell lines a useful model for host–pathogen–microbiome interaction studies.
3D Culture Models
An experimental system for host–microbe interactions based on 3D organoid cultures may act as a linkage between traditional 2D cell cultures and in vivo systems. Due to the relative simplicity of standard cell line models, many aspects of intestinal infections cannot be replicated in vitro. In 2009, Sato et al. (2009) identified essential growth factors for organoid culture and started an era of three-dimensional IEC systems in vitro. The advantage of these cultures over classical cell cultures includes better organization of cell junctions and an extracellular matrix and better expression of brush-border proteins. Organoids also possess crypt- and villus-like structures (Wilson et al., 2015) and therefore create a close-to-physiological environment for epithelial cells and microbiota (Mahe et al., 2013), providing a great opportunity for investigations focused on a three-way interaction. Unlike animal models, organoids allow the monitoring of host–microbiota interactions in a controlled environment. Furthermore, it provides the ability to work on human-derived cells and even precisely defined patient cells. Overall, the physiological relevance of the system makes organoids a promising technology for infectious diseases and microbiome studies.
Organoids can be derived from pluripotent stem cells or adult stem cells and can reproduce epithelial tissue in vitro (Barker et al., 2010). To support 3D growth, organoids are usually cultured in Matrigel, which provides chemical and mechanical support for intestinal stem cells, and cell proliferation is supported by numerous growth factors (Dutton et al., 2019). Organoids have great potential to be an ideal experimental model, as they are composed of patient-derived primary human epithelial cells, and are also accessible to experimental approaches. However, to date, despite numerous reports on organoids in microbiota investigation (reviewed in Bartfeld, 2016) as well as Salmonella studies using organoids (Zhang Y. G. et al., 2014; Yin and Zhou, 2018; Co et al., 2019), no study has used such a model for Salmonella–host–microbiota interaction. One possible reason for this is that only a few in vitro models allow the co-culture of a variety of bacteria with intestinal cells (reviewed in Elzinga et al., 2019). Nevertheless, the potential of the methods presented below creates the possibility that this model can also be efficiently applied in such experiments.
Previously, co-culture of intestinal organoids with microbiota, such as Lactobacillus, was demonstrated to have a positive impact on the barrier function of intestinal cells. L. rhamnosus enhances organoid proliferation and differentiation (Shaffiey et al., 2016), whereas Lactobacillus reuteri D8 stimulates the growth of intestinal organoids and protects against pro-inflammatory tumor necrosis factor α (TNF-α) (Hou et al., 2020). Microinjection of the non-pathogenic E. coli strain ECOR2 resulted in widespread transcriptional response and increased epithelial integrity (Hill et al., 2017). Further, short-chain fatty acids generated by Akkermansia muciniphila and Faecalibacterium prausnitzii strongly modulate transcription in mouse organoids (Arnold et al., 2016).
Organoids, as well as enteroids and colonoids, are reliable models for studying bacterial pathogenesis (Foulke-Abel et al., 2014; Zhang Y. G. et al., 2014). Many studies have used intestinal organoids to study interactions with enteric pathogens, such as Salmonella (Zhang Y. G. et al., 2014; Forbester et al., 2015; Wilson et al., 2015; Co et al., 2019). The complexity of organoid structure allows investigation of how pathogens interact with a variety of host cells and affect their function. For example, measurement of Salmonella entry into M-cells, stimulation of Paneth cells degranulation (Farin et al., 2014), or disruption of tight junctions (Zhang Y. G. et al., 2014) can be assessed. Both mice and human intestinal organoids infected with S. Typhimurium upregulate pro-inflammatory cytokine expression, such as interleukin-1β (IL-1β), TNF, and interleukin 8 (IL-8) (Zhang Y. G. et al., 2014; Forbester et al., 2015), and activate the nuclear factor kappa B (NF-κB) signaling pathway (Zhang Y. G. et al., 2014). However, Wilson et al. (2015) reported that mouse organoid cultures inhibit S. Typhimurium growth even 20 h post-infection by α-defensin activity. Interestingly, the ablation of matrix metalloproteinase 7 (Mmp7) in such organoids allows S. Typhimurium replication. The role of Peneth cells and secreted AMPs in the context of Salmonella and microbiota interaction was also intensively studied. Dysregulation of Peneth cells’ defensin secretion impacts the microbiome composition (Vaishnava et al., 2008), for example, by altering the balance between Firmicutes and Bacterioidates (Yokoi et al., 2019). At the same time, the absence of mice defensins (cryptdins) increases the host susceptibility to Salmonella infection (Salzman et al., 2003). Interestingly, the interaction of Salmonella with Peneth cells appears to be dependent on the serovar. It was shown that S. Typhimurium is able to enter mouse Paneth cells and induce their autophagy (Bel et al., 2017), whereas S. Enteritidis decreases autophagy, via the action of AvrA effector (Jiao et al., 2020).
The 3D structure of organoids mimics the organization of in vivo tissue, including the organization of both basal and apical sides outside and inside the spheroid, respectively (Figure 3). Most host–pathogen interactions using organoids involve microinjection of the bacterium inside the organoid structure. This allows the pathogen to reach its natural entry site and target apical tight junctions (McCracken et al., 2014; Bartfeld, 2016). Despite the technical problems related to precise microinjection, this method gives a chance to model the luminal antimicrobial response and investigate reactive oxygen species (Wilson et al., 2015, 2017). Another method for studying bacteria–organoid interactions is the dissociation of intestinal enteroids onto permeable transwell supports (VanDussen et al., 2015). This 2D culture method of 3D structures allows independent control of apical and basolateral surfaces and has been successfully used to study a co-culture model of enteroids and macrophages, ultimately enabling a thorough investigation of the innate immune response against enteric pathogens (Noel et al., 2017). The newest idea is to generate reverse polarity organoids, with apical surfaces exposed to the culture media and thus easily accessible for potential interactions (Co et al., 2019, 2021). Such inside-out enteroids open possibilities for studying host–pathogen–microbiota triangle in a more diverse and developed, but still relatively easy to control, environment. Organoids are one of the most promising models for host–pathogen–microbiota interactions (Table 2). The growth of the standardized, validated media and culture methods make this system a good balance between traditional 2D cell culture models and more complicated and less predictable animal models. The undeniable advantage of this system is its ability to investigate diverse human cells during infection by human pathogens.
Another interesting 3D model that may be useful for studying the host–pathogen–microbiome interaction is IVOC (Fang et al., 2013). It includes freshly obtained biopsies maintained in culture media under oxygenation to maintain the exchange of gas and nutrients. The material obtained by biopsy allows the study of the interaction of microbes with human tissue; however, it requires the delivery of fresh tissue from a clinic and generates variability in the samples due to different donors. IVOC allows the studies on the host–microbe interaction under physiologically relevant conditions, with the natural architecture of tissues and fully differentiated intestinal cells. Over the years, the traditional IVOC model (Knutton et al., 1984) was sequentially replaced by the polarized IVOC (El Asmar et al., 2002; Collins et al., 2010) with basal and apical sites available for interactions or by different variants adapted for other purposes (reviewed in Fang et al., 2013). Overall, this model provides a substantial advantage over cell cultures and is suitable for studying the interaction between pathogens and host cells (Haque et al., 2004; Schüller et al., 2009; Fang et al., 2013). What is more, it has also been applied to study probiotic bacteria impact on Salmonella infection (Collins et al., 2010).
Taken together, 3D infection models provide a unique opportunity to merge the study of a well-developed and diverse cellular mixture with a complexity close to in vivo experiments, with a relatively high controllability. From this perspective, most questions regarding host–pathogen–microbiome gameplay may be addressed using 3D models.
Organ-On-A-Chip
Most of the abovementioned models allow the co-culture of host intestinal cells and microbes for a few hours up to 1 day, which limits their use in long-term experiments. Some limitations can be avoided by using microfluidic models, in which multiple cell layers may be grown on thin chambers with channels divided by a thin membrane. These systems, classified as “organs-on-a-chip” are generated using soft lithography, a method similar to computer chip manufacturing. Currently, the use of multiple channel shapes, different intestinal cell types, fluid flow, and even vascular channels has become a promising model for in vitro studies. In addition, microchannels that support laminar fluid flow can mimic intestinal peristaltic movements, allowing a closer approximation of natural mechanical deformations as well as nutrient delivery and toxin removal. Flow shear stress generated by such models allows the development of a denser actin network, increased mitochondrial activity, metabolic enzyme expression, and gene expression profile at a level closer to that of the in vivo intestine (Delon et al., 2019). Moreover, spontaneous formation of villus structures and mucus production by epithelial cells can occur (Sung et al., 2011; Shim et al., 2017; Grassart et al., 2019), thereby providing a platform for creating niches for interactions of microbiota with pathogenic bacteria, including Salmonella (Costello et al., 2014). Unlike existing in vitro models, microfluid channels form complex interactions between aerobic bacteria, anaerobic bacteria, and host tissues. To apply microfluidic models to patient-specific content, intestinal chips with primary cell lines were introduced (Kasendra et al., 2018). These chips can better reproduce the normal epithelial physiology, allow the use of different intestinal cell types, and finally, allow to mimic the host response to bacterial infection and append the model for many possible applications, including inflammation and infection. The first microfluidic system to co-culture the Caco-2 cell line and L. rhamnosus for more than 1 week was presented by Kim et al. (2012) and was later replaced by a more sophisticated gut chip (Kim et al., 2016) and HuMix (human–microbial cross talk) models (Shah et al., 2016; Shin and Kim, 2018). A gut chip, which supports epithelial cells as well as capillary endothelium and immune cells, was successfully applied to the co-culture of Caco-2 cells with L. rhamnosus GG and enteroinvasive E. coli (Kim et al., 2016), demonstrating for the first time that this system can work in host–pathogen–microbiota interactions.
There are some concerns regarding the limited availability and reproducibility of the system between laboratories, mostly because the system is still in the developing stage. Nevertheless, organ-on-a-chip technology is a powerful tool for host–pathogen–microbiota interaction, and thanks to the introduction of flow that simulates peristaltic movements and allows toxin removal bring them closer to in vivo models but with the controllability higher than in vivo (Table 2).
Mice
Although salmonellosis outcomes can differ between species, laboratory mice as an experimental model have proven valuable in the development of human medicine. A remarkable advantage of mouse infection models over in vitro and ex vivo methods is their ability to allow for proper signaling in intestinal cells, polarized and organized microvilli, enormous heterogeneity of cells affected by a microbe, a complete set of AMPs, mucus production, and a natural environment for the microbiota. Notably, different strains of mice show different levels of susceptibility to Salmonella infection (Monack et al., 2004; Lawley et al., 2006), and the host genetic background can heavily influence the outcome of infection (Foster et al., 2021). Most research on Salmonella pathogenesis has historically used Salmonella-sensitive mice, such as BALB/c or BL-6 strains. These mice have a mutation in the Slc11a1 gene (also known as Nramp1), which makes them susceptible to systemic infection by Salmonella. The Slc11A1 mutation causes a defect in ion transport in phagocytic vesicles, which allows S. Typhimurium to survive in macrophages (Blackwell et al., 2001). This model mimics the infection of humans with the host-restricted serovar Typhi that causes typhoid fever; however, new humanized mouse models for S. Typhi has also been developed (Libby et al., 2010; Song et al., 2010). For non-typhoidal serovars, Slc11A1 mice strains possess a significant drawback. Even with a very low infection dose, they are rapidly affected within 10 days. Therefore, to study persistence, some researchers have used 129/SvJ or CBA mice that bear a functional Slc11A1 allele (Tsolis et al., 2011). S. Typhimurium persists in the GI tract for more than 30 days in these mice and has been found to persist in the mesenteric lymph nodes and gallbladder (Monack et al., 2004). Moreover, mouse strains with different Salmonella susceptibilities also differ in intestinal microbiome composition (Ferreira et al., 2011). For example, microbes overrepresented in 129S1/SvImJ mice, belonging to the phylum Bacteroidetes, are associated with better protection against inflammation but do not affect Salmonella colonization levels (Ferreira et al., 2011).
Colonization resistance provided by indigenous commensals is an important barrier to infection by enteric pathogens. Treatment with antibiotics, such as streptomycin, before infection has been shown to reduce the infectious dose of S. Typhimurium in mice by 100,000-fold and is therefore commonly used in some mouse models of Salmonella infection to circumvent colonization resistance (Barthel et al., 2003; Stecher et al., 2005). These mice have symptoms that are more similar to those of human gastroenteritis; however, in contrast to many other species, they do not exhibit massive luminal fluid secretion. The final effect of such treatments depends on the antibiotic type and dose (Ferreira et al., 2011). For example, microbiome disruption in C57BL/6 mice treated with various doses of streptomycin and vancomycin was more uniform for streptomycin treatment than for vancomycin treatment (Sekirov et al., 2008). Most antibiotic pretreatment studies utilize very high doses of streptomycin (20 mg/mouse) to completely disrupt the microbiota; however, even significantly lower doses, which do not affect the total number of intestinal microbes, make mice more susceptible to S. Typhimurium infection (Sekirov et al., 2008). Different antibiotics and different doses were also applied to the 129S1/SvImJ mouse model for persistent Salmonella infection (Ferreira et al., 2011). Researchers suggested that the microbiota composition of the 129S1/SvImJ strain is affected differently when treated with metronidazole and streptomycin compared to the C57BL/6 microbiome.
Antibiotic treatment also enables an additional opportunity—microbiota transplantation and therefore opens up new possibilities in Salmonella–host–microbiome interactions. To address the following, gnotobiotic mice are used as a mammalian model system where defined microbiomes can be used in a semi-controlled environment. Gnotobiotic mice possess a strictly controlled microbiota level and composition, starting from germ-free mice without microbial species, to mice colonized by a single microbial species, up to a small, precisely defined microbial population (Falk et al., 1998). The higher complexity of microbiota has been shown to increase protection against Salmonella-induced inflammation (Stecher et al., 2010). For example, a MET-1 defined microbiota population was generated as protection after antibiotic therapies (Martz et al., 2015). MET-1 did not affect S. Typhimurium levels in the colon but was found to decrease systemic colonization of C57BL/6 mice by Salmonella. Another example is low-complexity microbiota (LCM) mice, germ-free C57Bl/6 mice with altered Schaedler flora (ASF) (Stecher et al., 2010). In this case, microbiota composition is stable even over long periods, and the gut immune system is closer to normal compared to that of germ-free mice. Both gnotobiotic mice and antibiotic pretreatment models have been frequently used in investigations on the basics of colonization resistance generated by microbiota (Woo et al., 2008; Sekirov et al., 2010; Thiennimitr et al., 2011). Overall, mice lacking microbiota, either due to antibiotic treatment or germ-free, as well as mice with a LCM, with up to 20 species, do not show colonization resistance (Endt et al., 2010; Stecher et al., 2010). As streptomycin-pretreated mice do not develop diarrhea, novel one-day-old C57BL/6 mice have been suggested as a good model for Salmonella infection (Zhang K. et al., 2014). Moreover, it could be an interesting model from an interaction with the microbiota point of view. In addition to lacking fully developed intestine as well as M-cells, and possessing enterocytes as the major Salmonella entry site, neonatal mice do not possess an established intestinal microbiota even without antibiotic treatment.
In addition to colonization resistance (Endt et al., 2010), the intestinal microbiota plays a role in “pathogen clearance” (i.e., the elimination of the pathogen from intestinal niches after infection). After pathogen infection, even if the host fights out the invader, microbiota richness and diversity are usually strongly affected and must return to their original balance. During this re-growth process, microbiota species fight the pathogen, among others, by stimulating the mucosal immune system (Stecher et al., 2005).
Alterations in the microbiota richness and diversity caused by antibiotic treatment may lead to the generation of a “super-shedder” phenotype, characterized by severe mucosal inflammation, high loads of pathogens in the intestine, high levels of fecal shedding (more than 108/g of feces), and rapid transmission of Salmonella to naïve case mates (Lawley et al., 2008). The shedding level of individual mice, at least partially controlled by the microbiota, is a known phenomenon in the spread of infectious diseases (Lawley et al., 2008). Previously, it was shown that after streptomycin treatment of C57BL/6 mice, all animals became super-shedders within 24 h after S. Typhimurium infection (Barthel et al., 2003). The persistent infection model induces differentiation between non-super-shedder and super-shedder-specific microbiota, resulting in protection against antibiotic treatment in the latter (Gopinath et al., 2014). Despite the mouse model used in Salmonella–microbiota interaction experiments, additional complications arise. Unpredictable variations in microbiota populations not only between different mouse strains but also between isogenic mice supplied by different vendors or even by different time cohorts, strongly affect experimental outcomes (Stecher et al., 2010; Franklin and Ericsson, 2017; Velazquez et al., 2019). Therefore, any experiment regarding the microbiota should consider the aforementioned fluctuations.
Other Animal Models
In addition to the most abundant mice in vivo models, there are at least two animal models frequently used to investigate Salmonella–microbiota interactions—chickens and pigs. Chick and laying hens are useful models for Salmonella infections as poultry farms are severely affected by salmonellosis, and transmission from chicken eggs to humans is a leading cause of foodborne Salmonella outbreaks. However, infection with host-unrestricted serovars, such as S. Enteritidis, results in localized intestinal inflammation, without electrolyte efflux or diarrhea (Liu et al., 2018). Salmonella infection has been shown to impact chick microbiota composition and richness (Juricova et al., 2013; Liu et al., 2018; Menanteau et al., 2018; Kempf et al., 2020), especially when the first contact with the pathogen occurs in the 1st day after hatching.
Notably, the complexity of chicken microbiota increases significantly from day 1 to days 14–19 of life (Crhanova et al., 2011; Stanley et al., 2013; Liu et al., 2018; Khan and Chousalkar, 2020), and its composition depends solely on environmental factors. After day 1 of hatching, the microbiota complexity is usually very low and consists of approximately five different species (Crhanova et al., 2011). In the following days, microbiome complexity increases, reaching up to approximately 40 different species on day 19 (Crhanova et al., 2011; Stanley et al., 2013). Based on the heterogeneity of intestinal microbiota, infection of chicks with Salmonella at different time points after hatching leads to different infection outcomes and may shed a different light on the pathogen–microbiota interaction. Neonatal chicks with limited microbiota richness develop intestinal inflammation between 2 and 4 days post-infection with S. Enteritidis (Kogut et al., 2016), which is usually followed by systemic spread (Troxell et al., 2015; Litvak et al., 2019). Infection of young (1–4 days old) chicks with S. Enteritidis also contributes to delayed microbiota development and an overall reduction in richness and diversity (Juricova et al., 2013; Mon et al., 2015). Rapidly growing pathogens mainly reduced Clostridiales, Lactobacillales, and Bifidobacteriales just 3 days after infection (Juricova et al., 2013).
An interesting model was used by Barrow et al. (2015) in which the established microbiota from 40-week-old hens was transferred to neonatal chicks. This system was found to significantly reduce the variation in Salmonella susceptibility between 1-day-old chicks, and make them less susceptible overall. Another strategy that can be used to improve the resistance of neonatal chicks and their microbiota to wild-type Salmonella is the administration of attenuated Salmonella strains or probiotics (Azcarate-Peril et al., 2018). The last idea is based on competitive exclusion theory, which states that two closely related species cannot occupy the same ecological niche. It was first described in the chicken model by Rantala and Nurmi (1973), when the administration of attenuated S. Typhimurium to 1-day-old chicks competitively excludes the next dose of S. Typhimurium administered after 24 h. The positive effect of competitive exclusion in the reduction of Salmonella colonization was further confirmed using different Salmonella serovars and using many different experimental design (Barrow et al., 1987; Callaway et al., 2008; Crhanova et al., 2011).
Pigs are natural Salmonella hosts and, as frequently asymptotic carriers of many Salmonella serovars, act as a reservoir for human infection (Pires et al., 2017). Pig and human gastrointestinal tracts are very similar, allowing us to bypass the mice model to avoid colitis in the presence of their natural microbiota (Ahmer and Gunn, 2011; Zhang et al., 2013; Drumo et al., 2016). In addition, pigs infected with Salmonella develop an acute phase very early and shed a high concentration of Salmonella in the feces (Lynch et al., 2017). As in mice (Lawley et al., 2008) and chicks (Kempf et al., 2020) models, pigs can also develop “low-” and “high-shedder” phenotypes. Interestingly, a “low-” and “high-shedder” phenotypes differ significantly in the abundance of microbiota prior to Salmonella infection (Bearson et al., 2013; Argüello et al., 2019); this microbiota diversity may lead to the discovery of specific microbiota species and compositions, which may act as a marker for Salmonella susceptibility.
The age of potential hosts plays an important role in the selection of an appropriate pig model for our Salmonella–microbiota experiment. Interestingly, microbiome composition is very similar among naturally delivered neonates, and is not strongly affected by variation in host genetics or other environmental factors during the first few days of life (Inman et al., 2010). The complexity of the microbiome increases with age (Inoue et al., 2005), and evolves rapidly during GI tract development, shifting from Bacteroidetes to Firmicutes with age (Mach et al., 2015). Therefore, it may be interesting to study the impact of Salmonella on the unstable vs. stable microbiome in swine (reviewed in Gresse et al., 2017). Piglets can also be used as a model to determine the efficiency of probiotic strains against Salmonella infections. Administration of selected Lactobacillus strains results in reduced severity and lower pathogen loads after S. Typhimurium infection (Casey et al., 2007). However, weaned piglets supplemented with the well-characterized probiotic strain, Enterococcus faecium, and infected with S. Typhimurium did not have different clinical symptoms from the control pigs (Szabó et al., 2009). Bacterial loads in their feces and internal organs were even higher than those in the control. The composition of intestinal microbiota in piglets infected with S. Typhimurium was found to be strongly correlated with the infecting strain virulence (Drumo et al., 2016), and the wild-type strain significantly reduced SCFA-producing bacterial strains, such as Faecalibacterium, Roseburia, Butyrivibrio, and Clostridium genera. Of note, similar to the mouse model, Salmonella exploits inflammation to compete with the piglet microbiota (Chirullo et al., 2015).
Animal infection models, despite their lower level of controllability, bring substantial advantages to host–pathogen–microbiota investigations. In addition to the natural microbiome environment, appropriate physiological conditions, and a fully developed immune system, they provide a unique opportunity to study a dysbiosis process (Table 2). Most of the questions focused on Salmonella infection and its three-handed game with the host and the microbiota can be answered using animal models. Infected organisms possess their own natural microbiome, which can also be modified throughout the experiment. Indeed, this allows to involve all of the elements that may play a role in pathogen infection: (1) interaction of the host cells with the complete microbiome (bacteriome, virome, and mycobiome); (2) complex immune system reaction; (3) all available environmental niches; and (4) natural, appropriate physiological conditions for the microbiome and the pathogen.
On the other hand, there are at least two important drawbacks of animal models: ethical restrictions and low controllability of experimental variables.
Conclusion and Prospects
The traditional approach for investigating host–enteropathogens interactions is currently faced with a third player—trillions of microbes inhabiting GI tissues. In addition to the examination of pathogen gene/protein expression profiles and host responses, it is important to consider the impact of diverse, complicated bacterial, fungal, and viral populations that live in a specific symbiosis with the host.
A healthy and balanced intestinal microbiome provides human benefits to the host, whereas disruption of the microbiota favors pathogen infections. Enteric pathogens may also compete with the microbiome for nutrients and environmental niches. This dynamic, complicated, and sophisticated interaction force the necessity of rethinking the traditional host–pathogen interaction approach. Investigations covering the three-way gameplay between the host, its microbiota, and the infecting pathogen give a unique opportunity to deeply understand what is really happening during pathogenic infection.
Many of the different experimental models mentioned in this review have enabled us to investigate this complex interaction and ultimately add new data to the field of pathogen infection. To maintain the balance between costs and potential gain, the investigator’s needs and expectations should be precisely determined. It seems reasonable to start with cheap and relatively easy-to-use media flow models, followed by a creative combination of the aforementioned in vitro and in vivo models. The use of organoids and gut-on-a-chip may push in vitro interactions in the microbiota context to a higher complexity level. One step further is the genetically modified cell lines and animals with an enhanced or silenced expression of specific receptors, which may impact both microbiota and pathogen behavior. Another interesting idea is to investigate the molecular details at the single-cell level using high-throughput methods, which allow screening of entire microbial and host populations. Comparison of different models when addressing the experimental question may bring a complete, full set of information, starting with molecular events at the level of the particular cell and ending with the monitoring of animal infection from the perspective of populations. Finally, merging sophisticated bioinformatics methods with the traditional experimental part may shed new light on this host–pathogen–microbiota triangle of interactions.
Author Contributions
KG conceptualized, wrote the original draft and final version of the manuscript, visualized, and prepared the figures.
Funding
This work was supported by the Polish National Science Centre Grant by decision number DEC-2020/38/E/NZ6/00182 and co-financed under the Leading Research Groups support project from the subsidy increased for the period 2020–2025 in the amount of 2% of the subsidy referred to Art. 387 (3) of the Law of 20 July 2018 on Higher Education and Science, obtained in 2019.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adetoye, A., Pinloche, E., Adeniyi, B. A., and Ayeni, F. A. (2018). Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 18:96. doi: 10.1186/s12866-018-1248-y
Ahmer, B. M. M., and Gunn, J. S. (2011). Interaction of Salmonella spp. With the intestinal microbiota. Front. Microbiol. 2:101. doi: 10.3389/fmicb.2011.00101
Anderson, M. C., Vonaesch, P., Saffarian, A., Marteyn, B. S., and Sansonetti, P. J. (2017). Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21, 769–776e3. doi: 10.1016/j.chom.2017.05.004
Aoki-Yoshida, A., Saito, S., Fukiya, S., Aoki, R., Takayama, Y., Suzuki, C., et al. (2016). Lactobacillus rhamnosus GG increases toll-like receptor 3 gene expression in murine small intestine ex vivo and in vivo. Benef. Microbes 7, 421–429. doi: 10.3920/BM2015.0169
Argüello, H., Estellé, J., Leonard, F. C., Crispie, F., Cotter, P. D., O’Sullivan, O., et al. (2019). Influence of the intestinal microbiota on colonization resistance to Salmonella and the shedding pattern of naturally exposed pigs. mSystems 4:e00021-19. doi: 10.1128/mSystems.00021-19
Arnold, J. W., Roach, J., and Azcarate-Peril, M. A. (2016). Emerging technologies for gut microbiome research. Trends Microbiol. 24, 887–901. doi: 10.1016/j.tim.2016.06.008
Avendaño-Pérez, G., and Pin, C. (2013). Loss of culturability of salmonella enterica subsp. enterica serovar typhimurium upon cell-cell contact with human fecal bacteria. Appl. Environ. Microbiol. 79, 3257–3263. doi: 10.1128/AEM.00092-13
Avendaño-Pérez, G., Nueno-Palop, C., Narbad, A., George, S. M., Baranyi, J., and Pin, C. (2015). Interactions of Salmonella enterica subspecies enterica serovar Typhimurium with gut bacteria. Anaerobe 33, 90–97. doi: 10.1016/j.anaerobe.2015.02.006
Azcarate-Peril, M. A., Butz, N., Cadenas, M. B., Koci, M., Ballou, A., Mendoza, M., et al. (2018). An attenuated Salmonella enterica serovar Typhimurium strain and galactooligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl. Environ. Microbiol. 84:e02526-17. doi: 10.1128/AEM.02526-17
Barker, N., Huch, M., Kujala, P., van de Wetering, M., Snippert, H. J., van Es, J. H., et al. (2010). Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. doi: 10.1016/j.stem.2009.11.013
Barrow, P. A., Berchieri, A., Freitas Neto, O. C., and de Lovell, M. (2015). The contribution of aerobic and anaerobic respiration to intestinal colonization and virulence for Salmonella typhimurium in the chicken. Avian Pathol. 44, 401–407. doi: 10.1080/03079457.2015.1062841
Barrow, P. A., Huggins, M. B., Lovell, M. A., and Simpson, J. M. (1987). Observations on the pathogenesis of experimental Salmonella typhimurium infection in chickens. Res. Vet. Sci. 42, 194–199. doi: 10.1016/s0034-5288(18)30685-4
Bartfeld, S. (2016). Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev. Biol. 420, 262–270. doi: 10.1016/j.ydbio.2016.09.014
Barthel, M., Hapfelmeier, S., Quintanilla-Martínez, L., Kremer, M., Rohde, M., Hogardt, M., et al. (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003
Bearson, S. M. D., Allen, H. K., Bearson, B. L., Looft, T., Brunelle, B. W., Kich, J. D., et al. (2013). Profiling the gastrointestinal microbiota in response to Salmonella: low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 16, 330–340. doi: 10.1016/j.meegid.2013.03.022
Bel, S., Pendse, M., Wang, Y., Li, Y., Ruhn, K. A., Hassell, B., et al. (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. doi: 10.1126/science.aal4677
Bermudez-Brito, M., Muñoz-Quezada, S., Gomez-Llorente, C., Matencio, E., Bernal, M. J., Romero, F., et al. (2013). Cell-free culture supernatant of Bifidobacterium breve CNCM I-4035 decreases pro-inflammatory cytokines in human dendritic cells challenged with Salmonella typhi through TLR Activation. PLoS One 8:e59370. doi: 10.1371/journal.pone.0059370
Blackwell, J. M., Goswami, T., Evans, C. A. W., Sibthorpe, D., Papo, N., White, J. K., et al. (2001). SLC11A1 (formerly NRAMP1) and disease resistance. Cell. Microbiol. 3, 773–784. doi: 10.1046/j.1462-5822.2001.00150.x
Brosnahan, A. J., and Brown, D. R. (2012). Porcine IPEC-J2 intestinal epithelial cells in microbiological investigations. Vet. Microbiol. 156, 229–237. doi: 10.1016/j.vetmic.2011.10.017
Bucci, C., Santonicola, A., and Iovino, P. (2016). Gut microbiota and IBS 1 the normal microbiota: an essential factor for a healthy gut. Probiotics Prebiotics Synbiotics 557–566.
Burkholder, K. M., Fletcher, D. H., Gileau, L., and Kandolo, A. (2019). Lactic acid bacteria decrease Salmonella enterica Javiana virulence and modulate host inflammation during infection of an intestinal epithelial cell line. Pathog. Dis. 77:ftz025. doi: 10.1093/femspd/ftz025
Callaway, T. R., Edrington, T. S., Anderson, R. C., Harvey, R. B., Genovese, K. J., Kennedy, C. N., et al. (2008). Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Anim. Health Res. Rev. 9, 217–225. doi: 10.1017/S1466252308001540
Carey, C. M., and Kostrzynska, M. (2013). Lactic acid bacteria and bifidobacteria attenuate the proinflammatory response in intestinal epithelial cells induced by Salmonella enterica serovar typhimurium. Can. J. Microbiol. 59, 9–17. doi: 10.1139/cjm-2012-0446
Carman, R. J., and Woodburn, M. A. (2001). Effects of low levels of ciprofloxacin on a chemostat model of the human colonic microflora. Regul. Toxicol. Pharmacol. 33, 276–284. doi: 10.1006/rtph.2001.1473
Casey, P. G., Gardiner, G. E., Casey, G., Bradshaw, B., Lawlor, P. G., Lynch, P. B., et al. (2007). A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 73, 1858–1863. doi: 10.1128/AEM.01840-06
Chirullo, B., Pesciaroli, M., Drumo, R., Ruggeri, J., Razzuoli, E., Pistoia, C., et al. (2015). Salmonella typhimurium exploits inflammation to its own advantage in piglets. Front. Microbiol. 6:985. doi: 10.3389/fmicb.2015.00985
Cinquin, C., Le Blay, G., Fliss, I., and Lacroix, C. (2006a). Erratum: comparative effects of exopolysaccharides from lactic acid bacteria and fructo-oligosaccharides on infant gut microbiota tested in an in vitro colonic model with immobilized cells (FEMS Microbiology Ecology (2006) 57 (324-336)). FEMS Microbiol. Ecol. 57, 337–339. doi: 10.1111/j.1574-6941.2006.00118.x
Cinquin, C., Le Blay, G., Fliss, I., and Lacroix, C. (2006b). New three-stage in vitro model for infant colonic fermentation with immobilized fecal microbiota. FEMS Microbiol. Ecol. 57, 324–336. doi: 10.1111/j.1574-6941.2006.00117.x
Cleusix, V., Lacroix, C., Vollenweider, S., and Le Blay, G. (2008). Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiol. Ecol. 63, 56–64. doi: 10.1111/j.1574-6941.2007.00412.x
Co, J. Y., Margalef-Català, M., Li, X., Mah, A. T., Kuo, C. J., Monack, D. M., et al. (2019). Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509–2520.e4. doi: 10.1016/j.celrep.2019.01.108
Co, J. Y., Margalef-Català, M., Monack, D. M., and Amieva, M. R. (2021). Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat. Protoc. 16, 5171–5192. doi: 10.1038/s41596-021-00607-0
Collins, J. W., Coldham, N. G., Salguero, F. J., Cooley, W. A., Newell, W. R., Rastall, R. A., et al. (2010). Response of porcine intestinal in vitro organ culture tissues following exposure to Lactobacillus plantarum JC1 and Salmonella enterica serovar typhimurium SL1344. Appl. Environ. Microbiol. 76, 6645–6657. doi: 10.1128/AEM.03115-09
Costello, C. M., Sorna, R. M., Goh, Y. L., Cengic, I., Jain, N. K., and March, J. C. (2014). 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol. Pharm. 11, 2030–2039. doi: 10.1021/mp5001422
Crhanova, M., Hradecka, H., Faldynova, M., Matulova, M., Havlickova, H., Sisak, F., et al. (2011). Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar enteritidis infection. Infect. Immun. 79, 2755–2763. doi: 10.1128/IAI.01375-10
Crocker, P. R., and Feizit, T. (1996). Carbohydrate recognition systems: functional triads in cell-cell interactions. Curr. Opin. Struct. Biol. 6, 679–691. doi: 10.1016/s0959-440x(96)80036-4
Das, J. K., Mishra, D., Ray, P., Tripathy, P., Beuria, T. K., Singh, N., et al. (2013). In vitro evaluation of anti-infective activity of a Lactobacillus plantarum strain against Salmonella enterica serovar Enteritidis. Gut Pathog. 5:11. doi: 10.1186/1757-4749-5-11
de Masi, L., Yue, M., Hu, C., Rakov, A. V., Rankin, S. C., and Schifferli, D. M. (2017). Cooperation of adhesin alleles in Salmonella -host tropism. mSphere 2:e00066-17. doi: 10.1128/mSphere.00066-17
Delon, L. C., Guo, Z., Oszmiana, A., Chien, C. C., Gibson, R., Prestidge, C., et al. (2019). A systematic investigation of the effect of the fluid shear stress on Caco-2?cells towards the optimization of epithelial organ-on-chip models. Biomaterials 225:119521. doi: 10.1016/j.biomaterials.2019.119521
Dostal, A., Gagnon, M., Chassard, C., Zimmermann, M. B., O’Mahony, L., and Lacroix, C. (2014). Salmonella adhesion, invasion and cellular immune responses are differentially affected by iron concentrations in a combined in vitro gut fermentation-cell model. PLoS One 9:e93549. doi: 10.1371/journal.pone.0093549
Drumo, R., Pesciaroli, M., Ruggeri, J., Tarantino, M., Chirullo, B., Pistoia, C., et al. (2016). Salmonella enterica serovar typhimurium exploits inflammation to modify swine intestinal microbiota. Front. Cell. Infect. Microbiol. 5:106. doi: 10.3389/fcimb.2015.00106
Dutton, J. S., Hinman, S. S., Kim, R., Wang, Y., and Allbritton, N. L. (2019). Primary cell-derived intestinal models: recapitulating physiology. Trends Biotechnol. 37, 744–760. doi: 10.1016/j.tibtech.2018.12.001
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Microbiology: diversity of the human intestinal microbial flora. Science 308, 1635–1638.
El Asmar, R., Panigrahi, P., Bamford, P., Berti, I., Not, T., Coppa, G. V., et al. (2002). Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterology 123, 1607–1615. doi: 10.1053/gast.2002.36578
Elzinga, J., van der Oost, J., de Vos, W. M., and Smidt, H. (2019). The use of defined microbial communities to model host-microbe interactions in the human gut. Microbiol. Mol. Biol. Rev. 83:e00054-18. doi: 10.1128/MMBR.00054-18
Endt, K., Stecher, B., Chaffron, S., Slack, E., Tchitchek, N., Benecke, A., et al. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6:e1001097. doi: 10.1371/journal.ppat.1001097
Falk, P. G., Hooper, L. V., Midtvedt, T., and Gordon, J. I. (1998). Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62, 1157–1170. doi: 10.1128/MMBR.62.4.1157-1170.1998
Fang, S. B., Schüller, S., and Phillips, A. D. (2013). Human intestinal in vitro organ culture as a model for investigation of bacteria-host interactions. J. Exp. Clin. Med. 5, 43–50. doi: 10.1016/j.jecm.2013.02.006
Farin, H. F., Karthaus, W. R., Kujala, P., Rakhshandehroo, M., Schwank, G., Vries, R. G. J., et al. (2014). Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-γ. J. Exp. Med. 211, 1393–1340. doi: 10.1084/jem.20130753
Fattinger, S. A., Böck, D., Di Martino, M. L., Deuring, S., Ventayol, P. S., Ek, V., et al. (2020). Salmonella typhimurium discreet-invasion of the murine gut absorptive epithelium. PLoS Pathog. 16:e1008503. doi: 10.1371/journal.ppat.1008503
Fayol-Messaoudi, D., Berger, C. N., Coconnier-Polter, M. H., Liévin-Le Moal, V., and Servin, A. L. (2005). pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against Salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 71, 6008–6013. doi: 10.1128/AEM.71.10.6008-6013.2005
Fehlbaum, S., Chassard, C., Haug, M. C., Fourmestraux, C., Derrien, M., and Lacroix, C. (2015). Design and investigation of PolyFermS in vitro continuous fermentation models inoculated with immobilized fecal microbiota mimicking the elderly colon. PLoS One 10:e0142793. doi: 10.1371/journal.pone.0142793
Ferreira, R. B. R., Gill, N., Willing, B. P., Antunes, L. C. M., Russell, S. L., Croxen, M. A., et al. (2011). The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 6:e20338. doi: 10.1371/journal.pone.0020338
Forbester, J. L., Goulding, D., Vallier, L., Hannan, N., Hale, C., Pickard, D., et al. (2015). Interaction of Salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934. doi: 10.1128/IAI.00161-15
Foster, N., Tang, Y., Berchieri, A., Geng, S., Jiao, X., and Barrow, P. (2021). Revisiting persistent Salmonella infection and the carrier state: what do we know? Pathogens 10:1299. doi: 10.3390/pathogens10101299
Foulke-Abel, J., In, J., Kovbasnjuk, O., Zachos, N. C., Ettayebi, K., Blutt, S. E., et al. (2014). Human enteroids as an ex-vivo model of host–pathogen interactions in the gastrointestinal tract. Exp. Biol. Med. 239, 1124–1134. doi: 10.1177/1535370214529398
Franklin, C. L., and Ericsson, A. C. (2017). Microbiota and reproducibility of rodent models. Lab Anim. 46, 114–122. doi: 10.1038/laban.1222
Gahan, C. G. M., O’Sullivan, G. C., and Collins, J. K. (2011). “The gut microbiota, probiotics and infectious disease,” in Probiotic Bacteria and Enteric Infections: Cytoprotection by Probiotic Bacteria, eds J. J. Malago, J. F. J. G. Koninkx, and R. Marinsek-Logar (London: Springer).
Gieryńska, M., Szulc-Dąbrowska, L., Struzik, J., Mielcarska, M. B., and Gregorczyk-Zboroch, K. P. (2022). Integrity of the intestinal barrier: the involvement of epithelial cells and microbiota—a mutual relationship. Animals 12:145. doi: 10.3390/ani12020145
Gopinath, S., Lichtman, J. S., Bouley, D. M., Elias, J. E., and Monack, D. M. (2014). Role of disease-associated tolerance in infectious superspreaders. Proc. Natl. Acad. Sci. U.S.A. 111, 15780–15785. doi: 10.1073/pnas.1409968111
Grassart, A., Malardé, V., Gobba, S., Sartori-Rupp, A., Kerns, J., Karalis, K., et al. (2019). Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting Shigella infection. Cell Host Microbe 26, 435.e–444.e.
Gresse, R., Chaucheyras-Durand, F., Fleury, M. A., Van de Wiele, T., Forano, E., and Blanquet-Diot, S. (2017). Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. 25, 851–873. doi: 10.1016/j.tim.2017.05.004
Grzymajło, K., Ugorski, M., Kolenda, R., Kedzierska, A., Kuźmińska-Bajor, M., and Wieliczko, A. (2013). FimH adhesin from host unrestricted Salmonella enteritidis binds to different glycoprotein ligands expressed by enterocytes from sheep, pig and cattle than FimH adhesins from host restricted Salmonella Abortus-ovis, Salmonella choleraesuis and Salmonella. Vet. Microbiol. 166, 550–557. doi: 10.1016/j.vetmic.2013.07.004
Hao, S., Fan, Q., Bai, Y., Fang, H., Zhou, J., Fukuda, T., et al. (2020). Core fucosylation of intestinal epithelial cells protects against Salmonella typhi infection via up-regulating the biological antagonism of intestinal microbiota. Front. Microbiol. 11:1097. doi: 10.3389/fmicb.2020.01097
Haque, A., Bowe, F., Fitzhenry, R. J., Frankel, G., Thomson, M., Heuschkel, R., et al. (2004). Early interactions of Salmonella enterica serovar typhimurium with human small intestinal epithelial explants. Gut 53, 1424–1430. doi: 10.1136/gut.2003.037382
Hill, D. R., and Spence, J. R. (2017). Gastrointestinal organoids: understanding the molecular basis of the host–microbe interface. Cell Mol. Gastroenterol. Hepatol. 3, 138–149. doi: 10.1016/j.jcmgh.2016.11.007
Hill, D. R., Huang, S., Nagy, M. S., Yadagiri, V. K., Fields, C., Mukherjee, D., et al. (2017). Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. Elife 6:e29132. doi: 10.7554/eLife.29132
Höner zu Bentrup, K., Ramamurthy, R., Ott, C. M., Emami, K., Nelman-Gonzalez, M., Wilson, J. W., et al. (2006). Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes Infect. 8, 1813–1825. doi: 10.1016/j.micinf.2006.02.020
Hou, Q., Ye, L., Liu, H., Huang, L., Yang, Q., Turner, J. R., et al. (2020). Correction: Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22 (2018), 25, 9, (1657-1670), 10.1038/s4141. Cell Death Differ. 28, 2025–2027. doi: 10.1038/s41418-018-0070-2
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., Chinwalla, A. T., et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Inman, C. F., Haverson, K., Konstantinov, S. R., Jones, P. H., Harris, C., Smidt, H., et al. (2010). Rearing environment affects development of the immune system in neonates. Clin. Exp. Immunol. 160, 431–439. doi: 10.1111/j.1365-2249.2010.04090.x
Inoue, R., Tsukahara, T., Nakanishi, N., and Ushida, K. (2005). Development of the intestinal microbiota in the piglet. J. Gen. Appl. Microbiol. 51, 257–265. doi: 10.2323/jgam.51.257
Ito, T., Sekizuka, T., Kishi, N., Yamashita, A., and Kuroda, M. (2019). Conventional culture methods with commercially available media unveil the presence of novel culturable bacteria. Gut Microbes 10, 77–91. doi: 10.1080/19490976.2018.1491265
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Reddy, D. N. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Jiao, Y., Zhang, Y., Lin, Z., Lu, R., Xia, Y., Meng, C., et al. (2020). Salmonella enteritidis effector AvrA suppresses autophagy by reducing Beclin-1 protein. Front. Immunol. 11:686. doi: 10.3389/fimmu.2020.00686
Juricova, H., Videnska, P., Lukac, M., Faldynova, M., Babak, V., Havlickova, H., et al. (2013). Influence of Salmonella enterica serovar enteritidis infection on the development of the cecum microbiota in newly hatched chicks. Appl. Environ. Microbiol. 79, 745–747. doi: 10.1128/AEM.02628-12
Kanmani, P., and Kim, H. (2020). Beneficial effect of immunobiotic strains on attenuation of Salmonella induced inflammatory response in human intestinal epithelial cells. PLoS One 15:e0229647. doi: 10.1371/journal.pone.0229647
Kasendra, M., Tovaglieri, A., Sontheimer-Phelps, A., Jalili-Firoozinezhad, S., Bein, A., Chalkiadaki, A., et al. (2018). Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 8:2871. doi: 10.1038/s41598-018-21201-7
Kato, M. T., Field, J. A., and Lettinga, G. (1997). Anaerobe tolerance to oxygen and the potentials of anaerobic and aerobic cocultures for wastewater treatment. Braz. J. Chem. Eng. 14, 395–407. doi: 10.1590/s0104-66321997000400015
Kawarizadeh, A., Nojoomi, F., Tabatabaei, M., Hosseinzadeh, S., and Farzaneh, M. (2019). The effect of bacillus coagulans on cytotoxicity and apoptosis induced by Salmonella typhimurium in HT-29 cell culture. Iran. J. Microbiol. 11, 305–312.
Kawarizadeh, A., Pourmontaseri, M., Farzaneh, M., Hosseinzadeh, S., Ghaemi, M., Tabatabaei, M., et al. (2020). Interleukin-8 gene expression and apoptosis induced by Salmonella typhimurium in the presence of Bacillus probiotics in the epithelial cell. J. Appl. Microbiol. 131, 449–459. doi: 10.1111/jam.14898
Kawarizadeh, A., Pourmontaseri, M., Farzaneh, M., Hossinzadeh, S., and Pourmontaseri, Z. (2021). Cytotoxicity, apoptosis, and IL-8 gene expression induced by some foodborne pathogens in presence of Bacillus coagulans in HT-29 cells. Microb. Pathog. 150:104685. doi: 10.1016/j.micpath.2020.104685
Kempf, F., Menanteau, P., Rychlik, I., Kubasová, T., Trotereau, J., Virlogeux-Payant, I., et al. (2020). Gut microbiota composition before infection determines the Salmonella super- and low-shedder phenotypes in chicken. Microb. Biotechnol. 13, 1611–1630. doi: 10.1111/1751-7915.13621
Khailova, L., Mount Patrick, S. K., Arganbright, K. M., Halpern, M. D., Kinouchi, T., and Dvorak, B. (2010). Bifidobacterium bifidum reduces apoptosis in the intestinal epithelium in necrotizing enterocolitis. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G1118–G1127. doi: 10.1152/ajpgi.00131.2010
Khan, S., and Chousalkar, K. K. (2020). Short-term feeding of probiotics and synbiotics modulates caecal microbiota during Salmonella typhimurium infection but does not reduce shedding and invasion in chickens. Appl. Microbiol. Biotechnol. 104, 319–334. doi: 10.1007/s00253-019-10220-7
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174. doi: 10.1039/c2lc40074j
Kim, H. J., Li, H., Collins, J. J., and Ingber, D. E. (2016). Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U.S.Am. 113, E7–E15. doi: 10.1073/pnas.1522193112
Klasa, B., Kędzierska, A. E., and Grzymajło, K. (2020). Pre-growth culture conditions affect type 1 fimbriae-dependent adhesion of Salmonella. Int. J. Mol. Sci. 21:4206. doi: 10.3390/ijms21124206
Knutton, S., Lloyd, D. R., Candy, D. C. A., and McNeish, A. S. (1984). In vitro adhesion of enterotoxigenic Escherichia coli to human intestinal epithelial cells from mucosal biopsies. Infect. Immun. 44, 514–518. doi: 10.1128/iai.44.2.514-518.1984
Kogut, M. H., Swaggerty, C. L., Byrd, J. A., Selvaraj, R., and Arsenault, R. J. (2016). Chicken-specific kinome array reveals that Salmonella enterica serovar enteritidis modulates host immune signaling pathways in the cecum to establish a persistence infection. Int. J. Mol. Sci. 17:1207. doi: 10.3390/ijms17081207
Kolenda, R., Ugorski, M., and Grzymajlo, K. (2019). Everything you always wanted to know about Salmonella type 1 fimbriae, but were afraid to ask. Front. Microbiol. 10:1017. doi: 10.3389/fmicb.2019.01017
Lawley, T. D., Bouley, D. M., Hoy, Y. E., Gerke, C., Relman, D. A., and Monack, D. M. (2008). Host transmission of Salmonella enterica serovar typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 76, 403–416. doi: 10.1128/IAI.01189-07
Lawley, T. D., Chan, K., Thompson, L. J., Kim, C. C., Govoni, G. R., and Monack, D. M. (2006). Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. doi: 10.1371/journal.ppat.0020011
Le Blay, G., Rytka, J., Zihler, A., and Lacroix, C. (2009). New in vitro colonic fermentation model for Salmonella infection in the child gut. FEMS Microbiol. Ecol. 67, 198–207. doi: 10.1111/j.1574-6941.2008.00625.x
Lecuit, M., and Eloit, M. (2017). “The viruses of the gut microbiota,” in The Microbiota in Gastrointestinal Pathophysiology: Implications for Human Health, Prebiotics, Probiotics, and Dysbiosis, eds M. H. Floch, Y. Ringel, and W. A. Walker (Amsterdam: Elsevier).
Lesuffleur, T., Porchet, N., Aubert, J. P., Swallow, D., Gum, J. R., Kim, Y. S., et al. (1993). Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J. Cell Sci. 106(Pt 3), 771–783. doi: 10.1242/jcs.106.3.771
Leteurtre, E., Gouyer, V., Rousseau, K., Moreau, O., Barbat, A., Swallow, D., et al. (2004). Differential mucin expression in colon carcinoma HT-29 clones with variable resistance to 5-fluorouracil and methotrexate. Biol. Cell 96, 145–151. doi: 10.1016/j.biolcel.2003.12.005
Levine, U. Y., Bearson, S. M. D., and Stanton, T. B. (2012). Mitsuokella jalaludinii inhibits growth of Salmonella enterica serovar typhimurium. Vet. Microbiol. 159, 115–122. doi: 10.1016/j.vetmic.2012.03.027
Libby, S. J., Brehm, M. A., Greiner, D. L., Shultz, L. D., McClelland, M., Smith, K. D., et al. (2010). Humanized nonobese diabetic-scid IL2rγnull mice are susceptible to lethal Salmonella typhi infection. Proc. Natl. Acad. Sci. U.S.A. 107, 15589–15594. doi: 10.1073/pnas.1005566107
Litvak, Y., Mon, K. K. Z., Nguyen, H., Chanthavixay, G., Liou, M., Velazquez, E. M., et al. (2019). Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25, 128.e–139.e. doi: 10.1016/j.chom.2018.12.003
Liu, L., Lin, L., Zheng, L., Tang, H., Fan, X., Xue, N., et al. (2018). Cecal microbiome profile altered by Salmonella enterica, serovar enteritidis inoculation in chicken. Gut Pathog. 10:34. doi: 10.1186/s13099-018-0261-x
Liu, Y., Fatheree, N. Y., Mangalat, N., and Rhoads, J. M. (2010). Human-derived probiotic Lactobacillus reuteri strains differentially reduce intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G1087–G1096. doi: 10.1152/ajpgi.00124.2010
Lynch, H., Arguëllo, H., Walia, K., Lawlor, P. G., Duffy, G., Gardiner, G. E., et al. (2017). Evaluation of an alternative experimental infection method, which closely mimics the natural route of transmission of monophasic Salmonella typhimurium in pigs. Foodborne Pathog. Dis. 14, 23–28. doi: 10.1089/fpd.2016.2193
Ma, J., Zhang, Y., Xia, Y., and Sun, J. (2010). The inflammatory cytokine tumor necrosis factor modulates the expression of Salmonella typhimurium effector proteins. J. Inflamm. 7:42. doi: 10.1186/1476-9255-7-42
Macfarlane, G. T., Macfarlane, S., and Gibson, G. R. (1998). Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35, 180–187. doi: 10.1007/s002489900072
Mach, N., Berri, M., Estellé, J., Levenez, F., Lemonnier, G., Denis, C., et al. (2015). Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 7, 554–569. doi: 10.1111/1758-2229.12285
Mahe, M. M., Aihara, E., Schumacher, M. A., Zavros, Y., Montrose, M. H., Helmrath, M. A., et al. (2013). Establishment of gastrointestinal epithelial organoids. Curr. Protoc. Mouse Biol. 3, 217–240. doi: 10.1002/9780470942390.mo130179
Malik-Kale, P., Jolly, C. E., Lathrop, S., Winfree, S., Luterbach, C., and Steele-Mortimer, O. (2011). Salmonella- at home in the host cell. Front. Microbiol. 2:125. doi: 10.3389/fmicb.2011.00125
Martz, S. L. E., Mcdonald, J. A. K., Sun, J., Zhang, Y. G., Gloor, G. B., Noordhof, C., et al. (2015). Administration of defined microbiota is protective in a murine Salmonella infection model. Sci. Rep. 5:16094. doi: 10.1038/srep16094
McCracken, K. W., Catá, E. M., Crawford, C. M., Sinagoga, K. L., Schumacher, M., Rockich, B. E., et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. doi: 10.1038/nature13863
Menanteau, P., Kempf, F., Trotereau, J., Virlogeux-Payant, I., Gitton, E., Dalifard, J., et al. (2018). Role of systemic infection, cross contaminations and super-shedders in Salmonella carrier state in chicken. Environ. Microbiol. 20, 3246–3260. doi: 10.1111/1462-2920.14294
Mon, K. K. Z., Saelao, P., Halstead, M. M., Chanthavixay, G., Chang, H. C., Garas, L., et al. (2015). Salmonella enterica serovars enteritidis infection alters the indigenous microbiota diversity in young layer chicks. Front. Vet. Sci. 2:61. doi: 10.3389/fvets.2015.00061
Monack, D. M., Bouley, D. M., and Falkow, S. (2004). Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J. Exp. Med. 199, 231–241. doi: 10.1084/jem.20031319
Moshiri, M., Dallal, M. M. S., Rezaei, F., Douraghi, M., Sharifi, L., Noroozbabaei, Z., et al. (2017). The effect of Lactobacillus acidophilus PTCC 1643 on cultured intestinal epithelial cells infected with Salmonella enterica serovar enteritidis. Osong Public Health Res. Perspect. 8, 54–60. doi: 10.24171/j.phrp.2017.8.1.07
Naimi, S., Zirah, S., Ben, T. M., Theolier, J., Fernandez, B., Rebuffat, S. F., et al. (2020). Microcin J25 exhibits inhibitory activity against Salmonella newport in continuous fermentation model mimicking swine colonic conditions. Front. Microbiol. 11:988. doi: 10.3389/fmicb.2020.00988
Noel, G., Baetz, N. W., Staab, J. F., Donowitz, M., Kovbasnjuk, O., Pasetti, M. F., et al. (2017). A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 7:45270.
Park, S. H., Kim, S. A., Lee, S. I., Rubinelli, P. M., Roto, S. M., Pavlidis, H. O., et al. (2017). Original XPC™ effect on Salmonella typhimurium and cecal microbiota from three different ages of broiler chickens when incubated in an anaerobic in vitro culture system. Front. Microbiol. 8:1070. doi: 10.3389/fmicb.2017.01070
Payne, S., Gibson, G., Wynne, A., Hudspith, B., Brostoff, J., and Tuohy, K. (2003). In vitro studies on colonization resistance of the human gut microbiota to Candida albicans and the effects of tetracycline and Lactobacillus plantarum LPK. Curr. Issues Intest. Microbiol. 4, 1–8.
Pérez, J. C. (2021). Fungi of the human gut microbiota: roles and significance. Int. J. Med. Microbiol. 311:151490. doi: 10.1016/j.ijmm.2021.151490
Pires, S. M., de Knegt, L., and Hald, T. (2017). Estimation of the relative contribution of different food and animal sources to human Salmonella infections in the European Union. EFSA Support. Publ. 8:184E.
Poeker, S. A., Lacroix, C., De Wouters, T., Spalinger, M. R., Scharl, M., and Geirnaert, A. (2019). Stepwise development of an in vitro continuous fermentation model for the murine caecal microbiota. Front. Microbiol. 10:e0142793.
Rantala, M., and Nurmi, E. (1973). Prevention of the growth of Salmonella infantis in chicks by the flora of the alimentary tract of chickens. Br. Poult. Sci. 14, 627–630. doi: 10.1080/00071667308416073
Rescigno, M., Urbano, M., Valzasina, B., Francolini, M., Rotta, G., Bonasio, R., et al. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367. doi: 10.1038/86373
Rettedal, E. A., Gumpert, H., and Sommer, M. O. A. (2014). Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat. Commun. 5:4714. doi: 10.1038/ncomms5714
Rogers, A. W. L., Tsolis, R. M., and Bäumler, A. J. (2021). Salmonella versus the microbiome. Microbiol. Mol. Biol. Rev. 85:e00027-19. doi: 10.1128/MMBR.00027-19
Roto, S. M., Rubinelli, P. M., and Ricke, S. C. (2015). An introduction to the avian gut microbiota and the effects of yeast-based prebiotic-type compounds as potential feed additives. Front. Vet. Sci. 2:28. doi: 10.3389/fvets.2015.00028
Saini, S., Pearl, J. A., and Rao, C. V. (2009). Role of FimW, FimY, and FimZ in regulating the expression of type I fimbriae in Salmonella enterica serovar typhimurium. J. Bacteriol. 191, 3003–3010. doi: 10.1128/JB.01694-08
Salzman, N. H., Chou, M. M., de Jong, H., Liu, L., Porter, E. M., and Paterson, Y. (2003). Enteric Salmonella Infection inhibits paneth cell antimicrobial peptide expression. Infect. Immun. 71, 1109–1115. doi: 10.1128/IAI.71.3.1109-1115.2003
Salzman, N. H., Hung, K., Haribhai, D., Chu, H., Karlsson-Sjöberg, J., Amir, E., et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–82. doi: 10.1038/ni.1825
Salzman, N. H., Underwood, M. A., and Bevins, C. L. (2007). Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 19, 70–83. doi: 10.1016/j.smim.2007.04.002
Sana, T. G., Flaugnatti, N., Lugo, K. A., Lam, L. H., Jacobson, A., Baylot, V., et al. (2016). Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U.S.A. 113, E5044–E5051. doi: 10.1073/pnas.1608858113
Sassone-Corsi, M., and Raffatellu, M. (2015). No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 194, 4081–4087. doi: 10.4049/jimmunol.1403169
Sato, T., Vries, R. G., Snippert, H. J., Van De Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi: 10.1038/nature07935
Schierack, P., Kleta, S., Tedin, K., Babila, J. T., Oswald, S., Oelschlaeger, T. A., et al. (2011). E coli Nissle 1917 affects Salmonella adhesion to porcine intestinal epithelial cells. PLoS One 6:e14712. doi: 10.1371/journal.pone.0014712
Schierack, P., Nordhoff, M., Pollmann, M., Weyrauch, K. D., Amasheh, S., Lodemann, U., et al. (2006). Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 125, 293–305. doi: 10.1007/s00418-005-0067-z
Schüller, S., Lucas, M., Kaper, J. B., Girón, J. A., and Phillips, A. D. (2009). The ex vivo response of human intestinal mucosa to enteropathogenic Escherichia coli infection. Cell. Microbiol. 11, 521–530. doi: 10.1111/j.1462-5822.2008.01275.x
Sekirov, I., Gill, N., Jogova, M., Tam, N., Robertson, M., de Llanos, R., et al. (2010). Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 1, 30–41. doi: 10.4161/gmic.1.1.10950
Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y., Lupp, C., et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736. doi: 10.1128/IAI.00319-08
Shaffiey, S. A., Jia, H., Keane, T., Costello, C., Wasserman, D., Quidgley, M., et al. (2016). Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen. Med. 11, 45–61. doi: 10.2217/rme.15.70
Shah, P., Fritz, J. V., Glaab, E., Desai, M. S., Greenhalgh, K., Frachet, A., et al. (2016). A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 7:11535. doi: 10.1038/ncomms11535
Shim, K. Y., Lee, D., Han, J., Nguyen, N. T., Park, S., and Sung, J. H. (2017). Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 19:37. doi: 10.1007/s10544-017-0179-y
Shin, W., and Kim, H. J. (2018). Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. U.S.A. 115, E10539–E10547. doi: 10.1073/pnas.1810819115
Shkoporov, A. N., and Hill, C. (2019). Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25, 195–209. doi: 10.1016/j.chom.2019.01.017
Skjolaas, K. A., Burkey, T. E., Dritz, S. S., and Minton, J. E. (2007). Effects of Salmonella enterica serovar typhimurium, or serovar Choleraesuis, Lactobacillus reuteri and Bacillus licheniformis on chemokine and cytokine expression in the swine jejunal epithelial cell line, IPEC-J2. Vet. Immunol. Immunopathol. 115, 299–308. doi: 10.1016/j.vetimm.2006.10.012
Song, J., Willinger, T., Rongvaux, A., Eynon, E. E., Stevens, S., Manz, M. G., et al. (2010). A mouse model for the human pathogen salmonella typhi. Cell Host Microbe 8, 369–376. doi: 10.1016/j.chom.2010.09.003
Spoetti, T., Hausmann, M., Menzel, K., Piberger, H., Herfarth, H., Schoelmerich, J., et al. (2007). Role of soluble factors and three-dimensional culture in in vitro differentation of intestinal macrophages. World J. Gastroenterol. 13, 1032–1041. doi: 10.3748/wjg.v13.i7.1032
Stanley, D., Geier, M. S., Hughes, R. J., Denman, S. E., and Moore, R. J. (2013). Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One 8:e84290. doi: 10.1371/journal.pone.0084290
Stecher, B., Chaffron, S., Käppeli, R., Hapfelmeier, S., Freedrich, S., Weber, T. C., et al. (2010). Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6:e1000711. doi: 10.1371/journal.ppat.1000711
Stecher, B., Macpherson, A. J., Hapfelmeier, S., Kremer, M., Stallmach, T., and Hardt, W. D. (2005). Comparison of Salmonella enterica serovar typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect. Immun. 73, 3228–3241. doi: 10.1128/IAI.73.6.3228-3241.2005
Sung, J. H., Yu, J., Luo, D., Shuler, M. L., and March, J. C. (2011). Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 11, 389–392. doi: 10.1039/c0lc00273a
Szabó, I., Wieler, L. H., Tedin, K., Scharek-Tedin, L., Taras, D., Hensel, A., et al. (2009). Influence of a probiotic strain of Enterococcus faecium on Salmonella enterica serovar typhimurium DT104 infection in a porcine animal infection model. Appl. Environ. Microbiol. 75, 2621–2628. doi: 10.1128/AEM.01515-08
Thielemann, N., Herz, M., Kurzai, O., and Martin, R. (2022). Analyzing the human gut mycobiome – a short guide for beginners. Comput. Struct. Biotechnol. J. 20, 608–614. doi: 10.1016/j.csbj.2022.01.008
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Torraca, V., Masud, S., Spaink, H. P., and Meijer, A. H. (2014). Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. DMM Dis. Models Mech. 7, 785–797. doi: 10.1242/dmm.015594
Troxell, B., Petri, N., Daron, C., Pereira, R., Mendoza, M., Hassan, H. M., et al. (2015). Poultry body temperature contributes to invasion control through reduced expression of Salmonella pathogenicity island 1 genes in Salmonella enterica serovars typhimurium and enteritidis. Appl. Environ. Microbiol. 81, 8192–8201. doi: 10.1128/AEM.02622-15
Tsolis, R. M., Xavier, M. N., Santos, R. L., and Bäumler, A. J. (2011). How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect. Immun. 79, 1806–1814. doi: 10.1128/IAI.01369-10
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810.
Tyrer, P. C., Bean, E. G., Ruth Foxwell, A., and Pavli, P. (2011). Effects of bacterial products on enterocyte-macrophage interactions in vitro. Biochem. Biophys. Res. Commun. 413, 336–341. doi: 10.1016/j.bbrc.2011.08.100
Uraipan, S., Brigidi, P., and Hongpattarakere, T. (2014). Antagonistic mechanisms of synbiosis between Lactobacillus plantarum CIF17AN2 and green banana starch in the proximal colon model challenged with Salmonella Typhimurium. Anaerobe 28, 44–53. doi: 10.1016/j.anaerobe.2014.05.002
Ushijima, T., and Seto, A. (1991). Selected faecal bacteria and nutrients essential for antagonism of Salmonella typhimurium in anaerobic continuous flow cultures. J. Med. Microbiol. 35, 111–117. doi: 10.1099/00222615-35-2-111
Uzzau, S., Brown, D. J., Wallis, T., Rubino, S., Leori, G., Bernard, S., et al. (2000). Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125, 229–255. doi: 10.1017/s0950268899004379
Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L., and Hooper, L v (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A. 105, 20858–20863. doi: 10.1073/pnas.0808723105
VanDussen, K. L., Marinshaw, J. M., Shaikh, N., Miyoshi, H., Moon, C., Tarr, P. I., et al. (2015). Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920. doi: 10.1136/gutjnl-2013-306651
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2010). Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 309, 1–7. doi: 10.1111/j.1574-6968.2010.02000.x
Vasquez, K. S., Shiver, A. L., and Huang, K. C. (2018). Cutting the gordian knot of the microbiota. Mol. Cell 70, 765–767. doi: 10.1016/j.molcel.2018.05.034
Velazquez, E. M., Nguyen, H., Heasley, K. T., Saechao, C. H., Gil, L. M., Rogers, A. W. L., et al. (2019). Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol. 4, 1057–1064. doi: 10.1038/s41564-019-0407-8
Wilson, S. S., Bromme, B. A., Holly, M. K., Wiens, M. E., Gounder, A. P., Sul, Y., et al. (2017). Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog. 13:e1006446. doi: 10.1371/journal.ppat.1006446
Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C., and Smith, J. G. (2015). A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 8, 352–361. doi: 10.1038/mi.2014.72
Woo, H., Okamoto, S., Guiney, D., Gunn, J. S., and Fierer, J. (2008). A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS One 3:e1603. doi: 10.1371/journal.pone.0001603
Wood, T. E., Aksoy, E., and Hachani, A. (2020). From welfare to warfare: the arbitration of host-microbiota interplay by the type VI secretion system. Front. Cell. Infect. Microbiol. 10:587948. doi: 10.3389/fcimb.2020.587948
Yeung, C. Y., Chiang Chiau, J. S., Chan, W. T., Bin, J. C., Cheng, M. L., Liu, H. L., et al. (2013). In vitro prevention of salmonella lipopolysaccharide-induced damages in epithelial barrier function by various lactobacillus strains. Gastroenterol. Res. Pract. 2013:973209. doi: 10.1155/2013/973209
Yin, Y., and Zhou, D. (2018). Organoid and enteroid modeling of Salmonella infection. Front. Cell. Infect. Microbiol. 8:102. doi: 10.3389/fcimb.2018.00102
Yokoi, Y., Nakamura, K., Yoneda, T., Kikuchi, M., Sugimoto, R., Shimizu, Y., et al. (2019). Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci. Rep. 9:2710. doi: 10.1038/s41598-019-39610-7
Yue, M., Han, X., de Masi, L., Zhu, C., Ma, X., Zhang, J., et al. (2015). Allelic variation contributes to bacterial host specificity. Nat. Commun. 6:8754.
Zhang, K., Dupont, A., Torow, N., Gohde, F., Leschner, S., Lienenklaus, S., et al. (2014). Age-dependent enterocyte invasion and microcolony formation by Salmonella. PLoS Pathog. 10:e1004385. doi: 10.1371/journal.ppat.1004385
Zhang, N., Ju, Z., and Zuo, T. (2018). Time for food: the impact of diet on gut microbiota and human health. Nutrition 51-52, 80–85. doi: 10.1016/j.nut.2017.12.005
Zhang, Q., Widmer, G., and Tzipori, S. (2013). A pig model of the human gastrointestinal tract. Gut Microbes 4, 193–200. doi: 10.4161/gmic.23867
Zhang, Y. G., Wu, S., Xia, Y., and Sun, J. (2014). Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol. Rep. 2:e12147. doi: 10.14814/phy2.12147
Keywords: Salmonella, microbiome, infection models, host-pathogen interaction, pathogen-microbiota interactions, organoids, mice infection
Citation: Grzymajlo K (2022) The Game for Three: Salmonella–Host–Microbiota Interaction Models. Front. Microbiol. 13:854112. doi: 10.3389/fmicb.2022.854112
Received: 13 January 2022; Accepted: 11 March 2022;
Published: 18 April 2022.
Edited by:
Jun Sun, The University of Illinois at Chicago, United StatesReviewed by:
Todd Riley Callaway, The University of Georgia, United StatesMin Yue, Zhejiang University, China
Yongguo Zhang, University of Illinois at Chicago, United States
Copyright © 2022 Grzymajlo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krzysztof Grzymajlo, Krzysztof.grzymajlo@upwr.edu.pl, orcid.org/0000-0002-1163-0679
 Krzysztof Grzymajlo
Krzysztof Grzymajlo